Advanced pharmaceutical bulletin. 10(4):638-647.
doi: 10.34172/apb.2020.077
Research Article
Transcriptome Changes in Colorectal Cancer Cells upon Treatment with Avicequinone B
Yanet Ocampo 1, #  , Daneiva Caro 1, #
, Daneiva Caro 1, #  , David Rivera 1, Jhoan Piermattey 2, Ricardo Gaitán 2, Luis A. Franco 1, *
, David Rivera 1, Jhoan Piermattey 2, Ricardo Gaitán 2, Luis A. Franco 1, * 
Author information:
1Biological Evaluation of Promising Substances Group, Department of Pharmaceutical Sciences, University of Cartagena, Carrera 50 No. 29-11, 130014, Cartagena, Colombia.
2Natural Products Group, Department of Pharmaceutical Sciences, University of Cartagena, Carrera 50 No. 29-11, 130014, Cartagena, Colombia.
# Both authors contributed equally to this paper.
Abstract
Purpose:
Naphtho[2,3-b]furan-4,9-dione (Avicequinone B), a natural naphthoquinone isolated from the mangrove tree Avicennia alba , is recognized as a valuable synthetic precursor with anti-proliferative effect. However, the molecular mechanism involved in its bioactivity has not been investigated. This study aimed to determine the selectivity of avicequinone B against cancer cells and the transcriptomic changes induced in colorectal cancer (CRC).
Methods: The cytotoxic effect against adenocarcinoma-derived cells or fibroblasts was evaluated using MTT assay. In addition, CRC cells were treated with avicequinone B in different settings to evaluate colony-forming ability, cell cycle progression, apoptosis/necrosis induction, and transcriptome response by RNA-seq.
Results: Avicequinone B effectively reduced the viability of breast, colorectal, and lung adenocarcinoma cells with IC50 lower than 10 μM, while fibroblasts were less affected. The induction of G2/M arrest and necrosis-like cell death were observed in avicequinone B-treated HT-29 cells. Furthermore, RNA-seq revealed 490 differentially expressed genes, highlighting the reduction of interferon stimulated genes and proliferative signaling pathways (JAK-STAT, MAPK, and PI3K-AKT), as well as the induction of ferroptosis and miR-21 expression.
Conclusion: In short, these results demonstrated the therapeutic potential of avicequinone B and paved the foundation for elucidating its mechanisms in the context of CRC.
Keywords: Avicequinone B, Colorectal cancer, RNA-sequencing, Interferon stimulated genes, Ferroptosis, miR-21
Copyright and License Information
© 2020 The Authors.
This is an Open Access article distributed under the terms of the Creative Commons Attribution (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, as long as the original authors and source are cited. No permission is required from the authors or the publishers.
Introduction
Mangrove ecosystems inhabit the intertidal regions of tropical coastlines and statuaries, typically between 30° north and south of the equator.1 Recently, mangroves have gain increasing attention due to their remarkable adaptability/tolerance toward hostile environmental conditions (i.e. high salinity, high temperature, muddy anaerobic soils, extreme tides, and strong winds) resulting in novel bioactive products which could be successfully used to treat human diseases, including cancer.1,2 However, research to exploit the potential of mangrove-derived metabolites is still limited.
Avicennia (Avicenniaceae) is the only mangrove genus that occurs throughout the world, comprising eight species with diverse ecological, phytochemical, and ethnomedicinal importance. Among them, A. alba , A. officinalis , and A. marina are considered highly valuable species due to their medicinal properties and chemical composition. In fact, several types of bioactive metabolites have been obtained from Avicennia , including flavonoids, tannins, terpenoids, fatty acids, and naphthoquinones.3 Outstandingly, this genus is documented as a rich source of naphthoquinones which are recognized by their antibacterial, antioxidant, chemopreventive, and anti-cancer activities.4-7 Among them, naphtho[2,3-b ]furan-4,9-dione (avicequinone B) and other structurally-related annulated furan derivatives such as naphtho[1,2-b ]furan-4,5-dione, have demonstrated outstanding cytotoxicity against cancer cells.8-11
Avicequinone B was first isolated by Ito et al6 in 2000 from the stem bark of A. alba , and later described by Jia et al12 in the leaves of A. marina. Interestingly, the bioactivity of avicequinone B was enthusiastically studied since 1994, when Koyanagi et al.13 proposed a reliable method for its chemical synthesis; at a time when it was consider an unnatural analogue or a mere precursor during the biosynthetic process. From then on, optimization of the preparation of avicequinone B has been a prolific field, since it is an important precursor for the synthesis of new annulated furan-naphthoquinone derivatives.14 In our ongoing study to optimize the structure of cytotoxic naphthoquinones against colorectal cancer (CRC), avicequinone B was identified as an effective inhibitor of HT-29 cells proliferation.15 In this work, we aimed to investigate the selectivity of avicequinone B towards adenocarcinoma cell lines, as well as the molecular mechanisms involved in its cytotoxicity using transcriptome sequencing. Our results provided evidence of several novel mechanisms correlated with the reduction of an inflammatory gene program, inhibition of proliferative signaling, and the induction of ferroptosis, to explain the role of avicequinone B as a promising anti-CRC drug.
Materials and Methods
Test compound
Avicequinone B (naphtho[2,3-b ]furan-4,9-dione) was synthesized as previously described by Acuña et al15 Analytical data confirmed that the purity of test compound was >99%. For bioassays, the compound was dissolved in dimethylsulfoxide (DMSO, Fisher Scientific, USA) and diluted in complete medium when needed.
Cell culture
Cell lines from colorectal adenocarcinoma (HT-29), lung carcinoma (A549), prostate adenocarcinoma (PC-3), and breast adenocarcinoma (MDA-MB-231) as well as normal dermal fibroblasts (PCS-201-012), normal human fetal lung fibroblast (MRC-5), and Mus musculus embryo fibroblasts (3T3-L1), were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured as detailed in supplementary methods.
Cell viability assay
Cytotoxicity of avicequinone B was evaluated using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay as described in supplementary methods.
Clonogenic assay
The ability of avicequinone B to suppress the replicative capacity of HT-29 and MRC-5 cells was confirmed using the clonogenic assay.16 Briefly, 750 cells/well were cultured into a 6-well plate for 6 h, and treated with test compound (0-16.4 μM) or vehicle (DMSO) for 48 h. Subsequently, the culture medium was replaced and the colonies were allowed to form for 7 days. Finally, colonies were fixed, stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA), and counted using a stereomicroscope (EZ4 HD, Leica Microsystems, Singapore).
Cell cycle analysis
HT-29 cells (2×105 cells/mL) were exposed to avicequinone B (8.20 μM) or vehicle (DMSO) for 48 h. Afterwards, the cells were harvested, washed with PBS, and fixed with cold 66% ethanol at 4°C overnight. Then, the cells were stained with propidium iodide (PI) and incubated with RNase A, according to the manufacturer’s instruction (kit ab139418; Abcam, Cambridge, UK). The effect of avicequinone B on cell cycle distribution was determined with flow cytometry (Dako, Beckman Coulter Inc., CA, USA).
Annexin V-SYTOX flow cytometry assay
Apoptosis/Necrosis of CRC cells was evaluated by means of the Annexin V-FITC Apoptosis Detection Kit (ab14086; Abcam, Cambridge, UK). Briefly, HT-29 cells (2×105 cells/mL) were treated with avicequinone B (8.20 μM) or vehicle for 48 h. Then, the cells were harvested and stained with Annexin V-FITC and SYTOX Green dye to measure the proportion of live, apoptotic, and necrotic cells with flow cytometry ((Dako, Beckman Coulter Inc., CA, USA) ).
Total RNA extraction and analysis
HT-29 CRC cells were treated for 48 h with avicequinone B (8.20 μM) and total RNA was extracted using the GeneJET™ RNA Purification kit (Thermo Fisher Scientific, Vilnius, Lithuania) as described by the manufacturer. The isolated RNA was analyzed at Corporación CorpoGen (Bogotá, Colombia). RNA purity and concentration were measured using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Qubit®2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), while RNA integrity was assessed using the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). Samples concentration varied between 11.4 and 97.6 ng/μL; 260/280 ratio were above 1.85; and RNA integrity number (RIN) value ranging from 4.2-9.0, with the majority of samples with RIN>7.
RNA-sequencing analysis
Library preparation and sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA). Total RNA (150-500 ng) was used for library construction with the TruSeq™ RNA LT Sample Preparation kit (Illumina Inc., San Diego, CA, USA), according to the manufacturer’s instructions. Subsequently, the validation of the enriched samples libraries was performed using the Qubit®dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA, USA) and the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). The product was a smear ranging in size from 461-680 bp. The libraries were normalized and pooled at equimolar concentration (2 nM). Cluster generation was performed using 5 pM of pooled normalized libraries on the cBOT (Illumina Inc., San Diego, CA, USA). Finally, sequencing was performed on the Illumina HiSeq 2500 for 300 cycles (2×150 bp, paired end run) (Illumina, Inc, California, USA). The bioinformatic analysis was performed as detailed in supplementary methods.
Quantitative real-time PCR (RT-qPCR)
To validate the RNA-sequencing analysis, six differentially expressed genes - DEGs (three up-regulated and three down-regulated) were selected for RT-qPCR, which was performed as described in the supplementary methods. Primers sequences are listed in Table S1.
Statistical Analysis
Where appropriate, data was represented as a mean ± standard error of the mean (SEM). Data was analyzed by student t test or one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test. Statistical significance was considered at P < 0.05.
Results and Discussion
Avicequinone B demonstrated a selective inhibitory activity against cancer cells
As we recently reported,15 avicequinone B (naphtho[2,3-b ]furan-4,9-dione, Figure 1A) demonstrated a potent cytotoxic activity against CRC cells (HT-29) with an inhibitory concentration 50 (IC50) of 8.20±0.06 μM. Similarly, test compound affected the viability of cells from breast (MDA-MB-231; IC50=6.43 ± 0.28 μM), lung (A549; IC50=2.71 ± 0.18 μM), and prostate adenocarcinomas (PC3; IC50=11.65 ± 1.54 μM) (Figure 1B). The selectivity of avicequinone B was confirmed using lung fibroblasts (MRC-5; IC50=21.80 ± 2.68 μM), dermal fibroblasts (PCS-201-012; IC50=9.12 ± 0.29 μM), as well as mouse 3T3-L1 fibroblasts (IC50=10.71 ± 0.89 μM). Since avicequinone B and its angular analogue (naphtho[1,2-b ] furan-4,5-dione) have been previously identified as potent cytotoxic agents against cells from breast (i.e. MCF7 and MDA-MB-231) and lung (i.e. A549, H460, and NCI-H460) adenocarcinoma8-11; we continued the pharmacological study of the test compound using CRC cells.
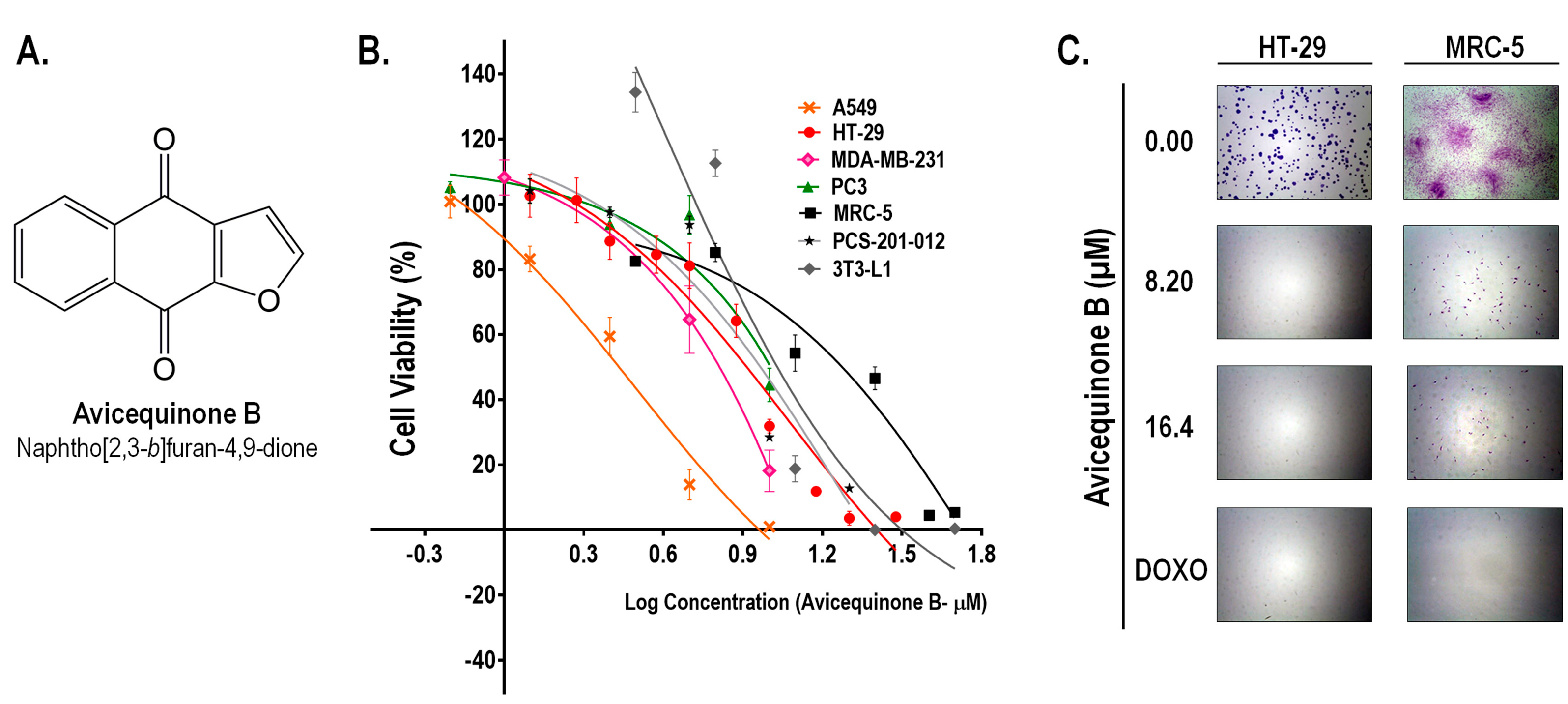
Figure 1.
Avicequinone B inhibits the viability and proliferation of cancer cells. (A) Chemical structure of the test compound. (B) The cell viability of several human adenocarcinoma cells (A549, HT-29, MDA-MB-231, and PC3) was efficiently and selectively reduced in comparison to normal fibroblasts (MRC5, PCS-201-012, and 3T3-L1). The percentage of viable cells in control was set as 100 %. Results are representative of at least two independent experiments and are expressed as the mean ± S.E.M. (n =6-11 per data group). (C) The colony forming-ability of HT-29 cells was completely suppressed by avicequinone B (8.20 and 16.40 µM) whereas human normal fibroblasts were partially affected. The chemotherapeutic drug, doxorubicin (DOXO), was used as reference. Representative pictures of colonies are shown (8X).
.
Avicequinone B inhibits the viability and proliferation of cancer cells. (A) Chemical structure of the test compound. (B) The cell viability of several human adenocarcinoma cells (A549, HT-29, MDA-MB-231, and PC3) was efficiently and selectively reduced in comparison to normal fibroblasts (MRC5, PCS-201-012, and 3T3-L1). The percentage of viable cells in control was set as 100 %. Results are representative of at least two independent experiments and are expressed as the mean ± S.E.M. (n =6-11 per data group). (C) The colony forming-ability of HT-29 cells was completely suppressed by avicequinone B (8.20 and 16.40 µM) whereas human normal fibroblasts were partially affected. The chemotherapeutic drug, doxorubicin (DOXO), was used as reference. Representative pictures of colonies are shown (8X).
Clonogenic assay showed that cytotoxic concentrations of avicequinone B (8.20 and 16.40 μM) suppressed the colony-forming ability of HT-29 cells, confirming its effectiveness against CRC cells (Figure 1C). In contrast, MRC-5 fibroblasts were alive and some colonies were observed; however the treatment with test compound significantly impaired their proliferative rate, raising concern about its safety. Therefore, to continue our work with avicequinone B as a potential lead compound to develop new anti-CRC drugs, a battery of three in vitro genotoxicity tests (Ames test, Micronucleus assay, and Comet test) was carried out as described in supplementary methods. Results evidenced the lack of mutagenicity of avicequinone B when employing its effective cytotoxic concentration (IC50 = 8.20 μM) (Figure s1).
Anti-proliferative effect of avicequinone B occurs by arresting cell cycle progression and inducing cellular death
As can be seen in Figure 2A, the treatment with avicequinone B, resulted in a significant increase in the proportion of 4N cells (G2/M phase) from 19.58±2.79% (control-0 μM) to 34.90±3.98% (avicequinone B-8.20 μM), whereas 2N (G1) and 2N-4N (S) cell populations tended to be reduced. Thus, avicequinone B triggers the arrest at the G2/M phase in HT-29 cells. Consistently, Tseng et al9 reported G2/M arrest when K562 leukemia cells were treated with this compound.
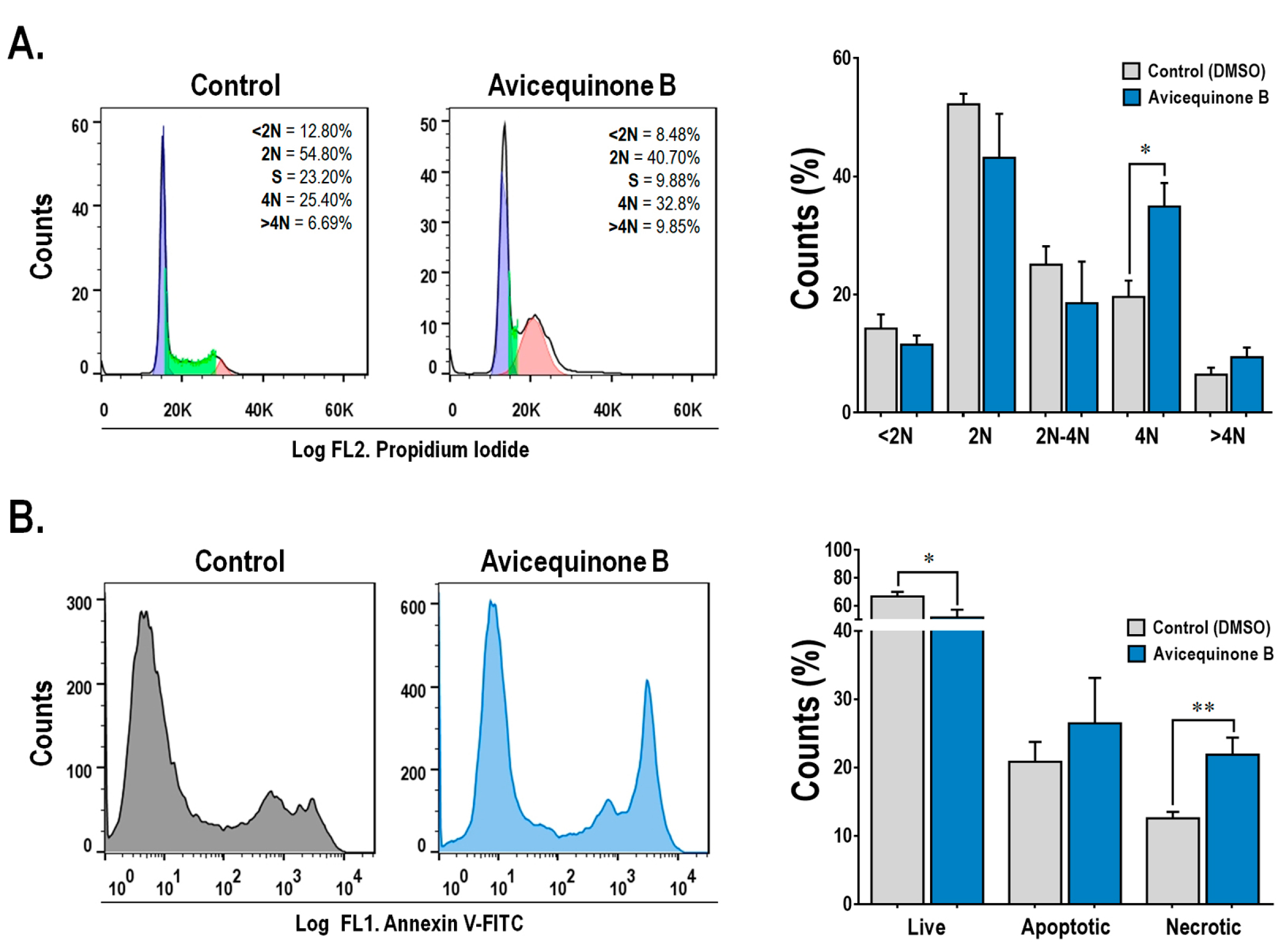
Figure 2.
Avicequinone B induces cell cycle arrest and necrosis-like cell death. Flow cytometry data showed that (A) HT-29 cells were accumulating at the G2/M phase (4N) and (B) undergoing cell death through necrosis, when treated with avicequinone B (8.20 µM) for 48 h. Distinctive histograms are presented showing the number of cells (vertical axis) vs. DNA content-propidium iodide or Annexin-V/Sytox green staining (horizontal axis-FL1 and FL2 channel, respectively). Results are representative of at least three independent experiments and are expressed as the mean ± S.E.M. (n=3-10 per group). Significant differences from the control are indicated by *P <0.05; **P <0.01 (ANOVA).
.
Avicequinone B induces cell cycle arrest and necrosis-like cell death. Flow cytometry data showed that (A) HT-29 cells were accumulating at the G2/M phase (4N) and (B) undergoing cell death through necrosis, when treated with avicequinone B (8.20 µM) for 48 h. Distinctive histograms are presented showing the number of cells (vertical axis) vs. DNA content-propidium iodide or Annexin-V/Sytox green staining (horizontal axis-FL1 and FL2 channel, respectively). Results are representative of at least three independent experiments and are expressed as the mean ± S.E.M. (n=3-10 per group). Significant differences from the control are indicated by *P <0.05; **P <0.01 (ANOVA).
On the other hand (Figure 2B), the staining with Annexin V-FITC and SYTOX demonstrated that avicequinone B (8.20 μM) enhanced the proportion of highly intense green fluorescence HT-29 cells (indicative of necrosis), without significantly increasing apoptotic cells (positive green fluorescence). This result differs from that obtained by Prateep et al8 who reported the induction of apoptosis, but not necrosis, in H460 cells treated with avicequinone B. This discrepancy might be explained by the differences in the experimental setting, since they used a mild test concentration (lower than the IC50).
Transcriptome sequencing revealed that avicequinone B promoted cellular death mainly by reducing the expression of IFN-stimulated genes.
To evaluate the mechanism involved in the bioactivity of avicequinone B against CRC, an RNA sequencing (RNA-seq)-based transcriptomic analysis was carried out using HT-29 cells treated with vehicle (control) or a toxic concentration of test compound (8.20 μM). The summary of the RNA-seq data showed similarity of reads, mapping rates, and GC content within the independent analyzed samples (Table S2, n = 3). In this experiment, 490 differentially expressed genes were identified out of a total of 12761 genes with measured expression (Figure 3A). These were obtained using a threshold of 0.05 for statistical significance (p- and FDR -q - values) and a log fold change of expression with absolute value of at least 0.5. Avicequinone B significantly down-regulated the expression of 341 genes while 149 genes were up-regulated (Table S3). The expression levels of a subset of DEGs, 3 up-regulated (SPRR1B, AKAP12, CYP1A1) and 3 down-regulated (IFITM1, IFI44L, EPSTI1), were measured via RT-qPCR to confirm the reliability of the transcriptomic analysis. The RT-qPCR results were consistent with those of the RNA-seq, showing a significant correlation (least-squares linear regression, P < 0.05; R2= 0.909-0.935; Figure S2), thus validating the analysis.
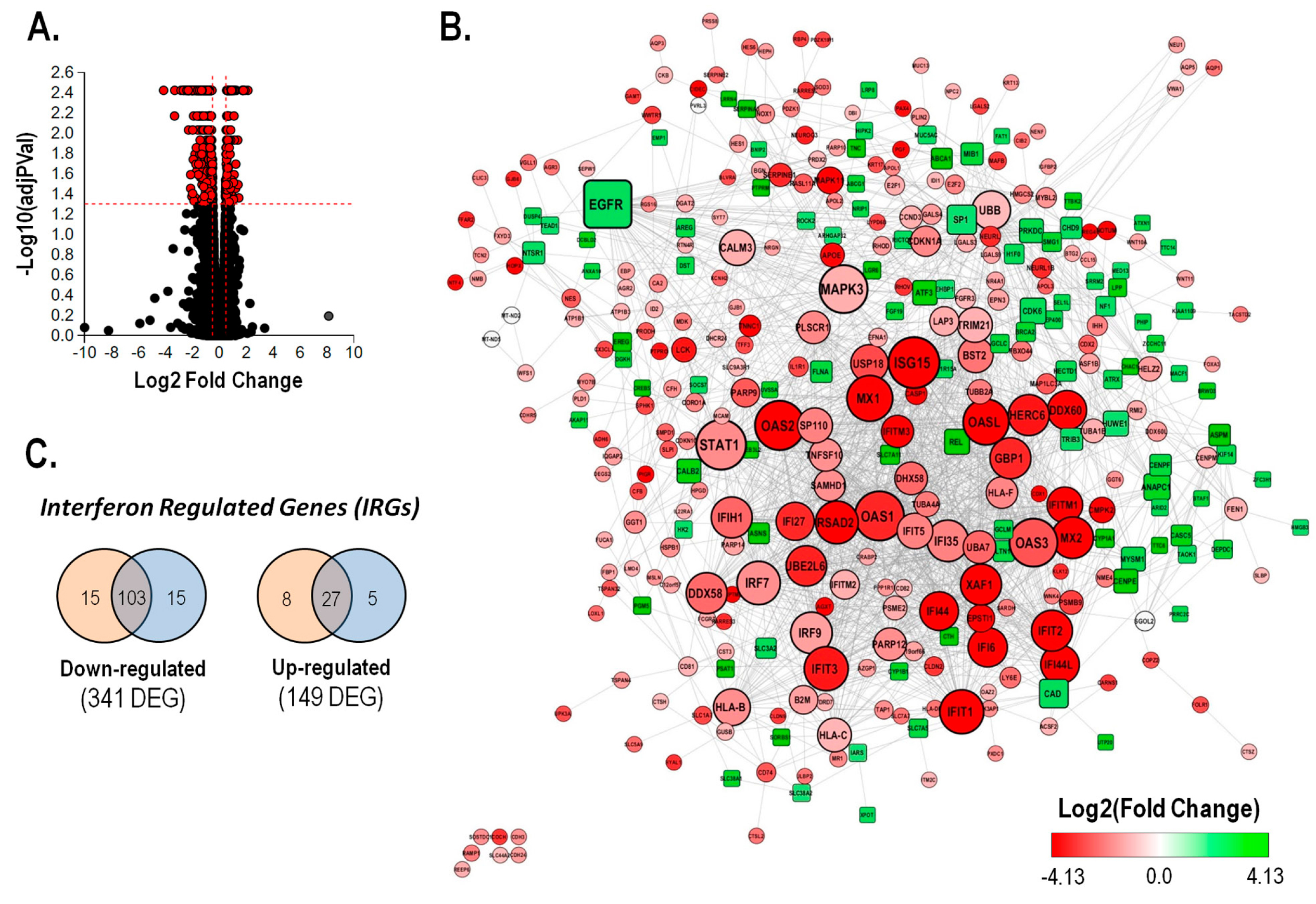

Figure 3.
Treatment with avicequinone B reduced the inflammatory transcriptome signature of HT-29 CRC cells by reducing the expression of interferon regulated genes.Transcriptomic analysis was performed by means of RNA-sequencing (A) Volcano plot presenting the mRNA expression changes of HT-29 cells treated with avicequinone B (8.20 μM) compared to control. All measured genes (dots) are represented in terms of their calculated expression change (x-axis) and the significance of the change (y-axis). The significance is represented in terms of the negative log (base 10) of the p -value, so that more significant genes are plotted higher on the y-axis. The dotted lines represent the thresholds used to select the differentially expressed genes (DEGs): 0.5 for expression change and 0.05 for significance. DEGs are presented as red dots (490). (B) The protein-protein interaction network of DEGs was built using STRING database at medium confidence (score=0.4) and analyzed using Cytoscape. Each node represents a gene/protein, the edge represents the interaction between gene/proteins, the size of the node is proportional to the degree of the node, the shape indicates the direction of the expression (square: up-regulation and circle: down-regulation), and the color represents the Log2 Fold Change. (C) Number of type I (orange) and type II (blue) interferon regulated genes (IRGs) that were identified in the list of DEGs regulated by avicequinone B using the Interferome database, filtering species (Homo sapiens ) and system (gastrointestinal tract).
.
Treatment with avicequinone B reduced the inflammatory transcriptome signature of HT-29 CRC cells by reducing the expression of interferon regulated genes.Transcriptomic analysis was performed by means of RNA-sequencing (A) Volcano plot presenting the mRNA expression changes of HT-29 cells treated with avicequinone B (8.20 μM) compared to control. All measured genes (dots) are represented in terms of their calculated expression change (x-axis) and the significance of the change (y-axis). The significance is represented in terms of the negative log (base 10) of the p -value, so that more significant genes are plotted higher on the y-axis. The dotted lines represent the thresholds used to select the differentially expressed genes (DEGs): 0.5 for expression change and 0.05 for significance. DEGs are presented as red dots (490). (B) The protein-protein interaction network of DEGs was built using STRING database at medium confidence (score=0.4) and analyzed using Cytoscape. Each node represents a gene/protein, the edge represents the interaction between gene/proteins, the size of the node is proportional to the degree of the node, the shape indicates the direction of the expression (square: up-regulation and circle: down-regulation), and the color represents the Log2 Fold Change. (C) Number of type I (orange) and type II (blue) interferon regulated genes (IRGs) that were identified in the list of DEGs regulated by avicequinone B using the Interferome database, filtering species (Homo sapiens ) and system (gastrointestinal tract).
The expression level of DEGs was integrated within a protein-protein interaction network built with STRING database (interaction score >0.4). Hub genes in the network were identified by topological analysis using the Cytoscape Network Analyzer. As shown in Figure 3B, most of the nodes with higher degree (ISG15, STAT1, OAS1, OAS2, OAS3, MAPK3, EGFR, MX1, OASL, and IFIT1) are interferon regulated genes (IRGs) whose expression was reduced by avicequinone B. Among these, STAT1, ISG15, MAPK3, and to a lesser extent OAS2 presented the highest centrality scores. Moreover, nearly 39% of the down-regulated DEGs are predicted to be regulated by type I and/or type II interferon (Figure 3C), as classified in the Interferome database. Likewise, out of the top 10 down-regulated genes (Table S4), seven were identified as IRGs (IFITM1, IFI44L, RSAD2, EPSTI1, TNNC1, GPRC5B, and CMPK2).
Simultaneously, the analysis of the top 10 up-regulated genes by avicequinone B (Table S5) showed that some were associated with the cornified envelope, epidermis development and cellular junction (SPRR1B, SCEL, and TNC); detoxification (CYP1A1), and tumor suppression in CRC (AKAP12). In addition, avicequinone B induced the expression of genes with low abundance mRNA in affected tissue or cells isolated from CRC patients (SLC7A11, SCEL, LRRN4, and SERPINA3). From this analysis, it was also noted that avicequinone B increased the expression of two markers of CRC metastasis: HOPX (down-regulated) and TNC (up-regulated), probably as a survival response of injured HT-29 cells.
Gene expression data was examined in the context of pathways, gene ontologies, and miRNAs, using the iPathway online software. As a result, 49 pathways were found to be significantly impacted. In addition, 1684 gene ontology (GO) terms and 5 microRNAs (miRNAs) were found to be significantly enriched based on uncorrected p -values. Figure 4A summarizes the significantly enriched GO terms related to biological process, molecular function, and cellular component, emphasizing the top-ten terms. Results demonstrated that avicequinone B induced a significant enrichment of terms related to the onset or progression of cancer such as cellular proliferation, cellular differentiation, cellular migration, angiogenesis, regulation of programmed cell death, and apoptotic process. Moreover, terms related to cell-cell signaling, cell-cell adhesion, assembly of the plasma membrane and extracellular space, response to drugs, and response to oxidative stress were also enriched. In particular, the treatment with avicequinone B promoted the differential expression of genes associated to immune response terms related to interferon (IFN) signaling, response to IFNs, and response to virus. Furthermore, the impact analysis revealed that among the top twenty-five influenced pathways (Figure 4B); eight are related to viral infections (influenza A, hepatitis C, human papillomavirus infection, herpes simplex infection, measles, Kaposi’s sarcoma-associated herpes virus infection, viral myocarditis, and HTLV-I infection). This distinctive association of avicequinone B with terms and pathways related to immune response to virus is explained by the remarkable down-regulation of IRGs, as described above.
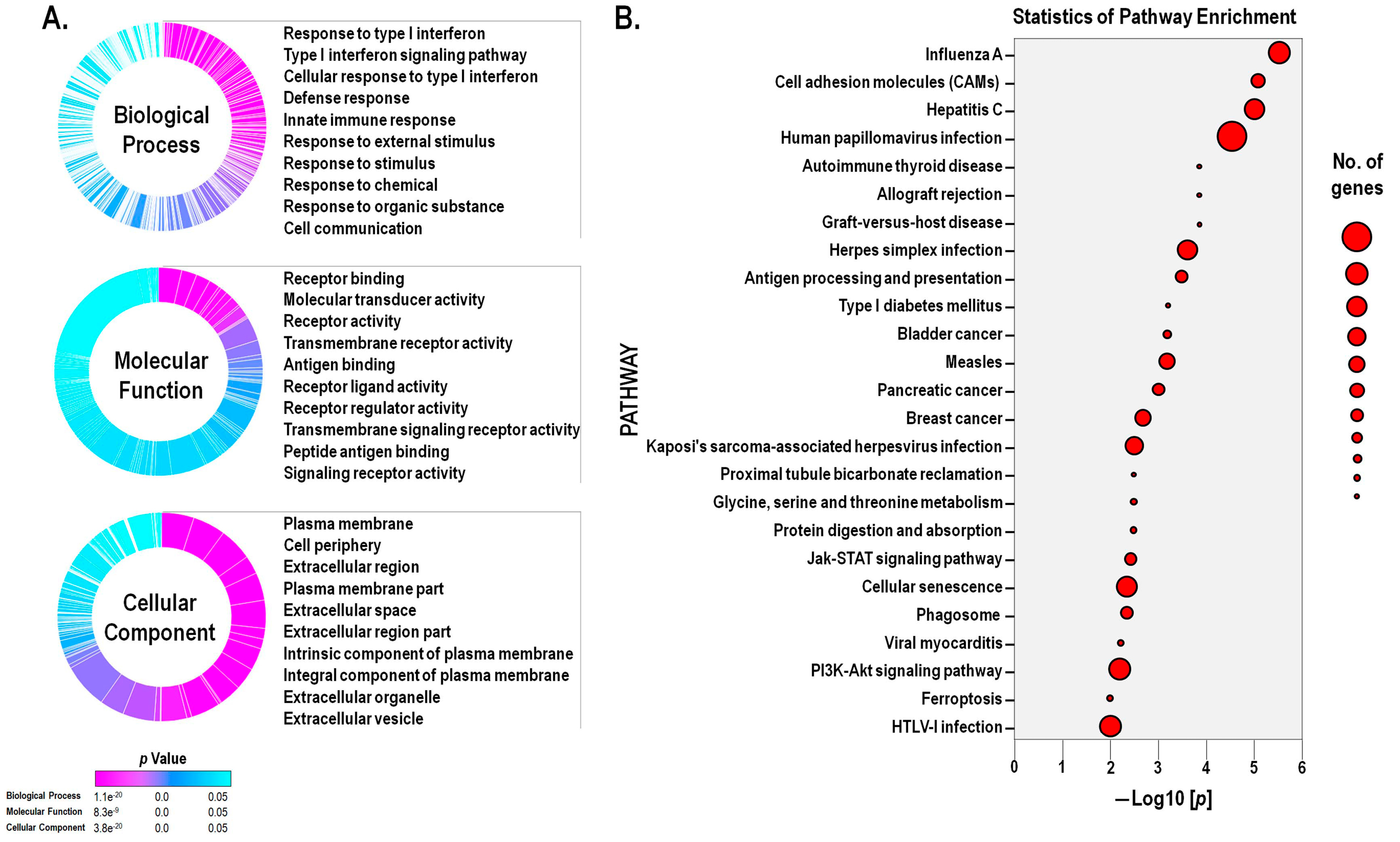

Figure 4.
Avicequinone B promoted alterations in the gene expression associated with cellular survival, proliferation, junction and signaling; response to interferon and virus; cancer pathways; and ferroptosis. (A) Differentially expressed genes (DEGs), between avicequinone B (8.20 µM)-treated HT-29 cells and control (vehicle), were categorized within three domains: biological processes, molecular function, and cellular components, through gene ontology (GO) term enrichment analysis performed with iPathwayGuide online software; cut-off p value p <0.05 corrected for multiple comparisons using FDR and Bonferroni. The GO summary graphs plot the significant overrepresented terms for each domain. Each section of the graph represents a different term, the height of the section is proportional to the number of DEGs, and the enrichment significance is displayed using a color scale with cyan being least significant and magenta most significant. (B) Scatter plot of the top twenty-five significantly enriched pathways for DEGs, as categorized by iPathwayGuide online software. Pathways are ranked (x-axis) according to their significance (y-axis) which is represented in terms of the negative log (base 10) of the P value (without correction). The size of the dots represents the number of DEGs.
.
Avicequinone B promoted alterations in the gene expression associated with cellular survival, proliferation, junction and signaling; response to interferon and virus; cancer pathways; and ferroptosis. (A) Differentially expressed genes (DEGs), between avicequinone B (8.20 µM)-treated HT-29 cells and control (vehicle), were categorized within three domains: biological processes, molecular function, and cellular components, through gene ontology (GO) term enrichment analysis performed with iPathwayGuide online software; cut-off p value p <0.05 corrected for multiple comparisons using FDR and Bonferroni. The GO summary graphs plot the significant overrepresented terms for each domain. Each section of the graph represents a different term, the height of the section is proportional to the number of DEGs, and the enrichment significance is displayed using a color scale with cyan being least significant and magenta most significant. (B) Scatter plot of the top twenty-five significantly enriched pathways for DEGs, as categorized by iPathwayGuide online software. Pathways are ranked (x-axis) according to their significance (y-axis) which is represented in terms of the negative log (base 10) of the P value (without correction). The size of the dots represents the number of DEGs.
Additionally, avicequinone B significantly impacted five cancer pathways -including bladder cancer (KEGG: 05219), pancreatic cancer (KEGG: 05212), and breast cancer (KEGG: 05224) (Figure S4)- displaying a shared differential expression of genes related to cellular proliferation, differentiation and/or migration (ERK and EGFR), as well as cell cycle progression (p21 and E2F). Interestingly, the test compound down-regulated oncogenes (FGFR3 and HES1) and genes belonging to the WNT family, while up-regulated tumor suppressor genes (BRCA2). Gene pathway analysis also revealed that avicequinone B impacted pathways related to development and homeostasis (JAK-STAT signaling pathway-KEGG: 04630) (Figure 5A); regulated cell death (ferroptosis- KEGG: 04216) (Figure 5B); cellular interactions (cellular adhesion molecules-KEGG: 04514); cellular proliferation and survival (PI3K-AKT signaling pathway-KEGG: 04151; and MAPK signaling pathway-KEGG: 04010) (Figure S5 and S6).
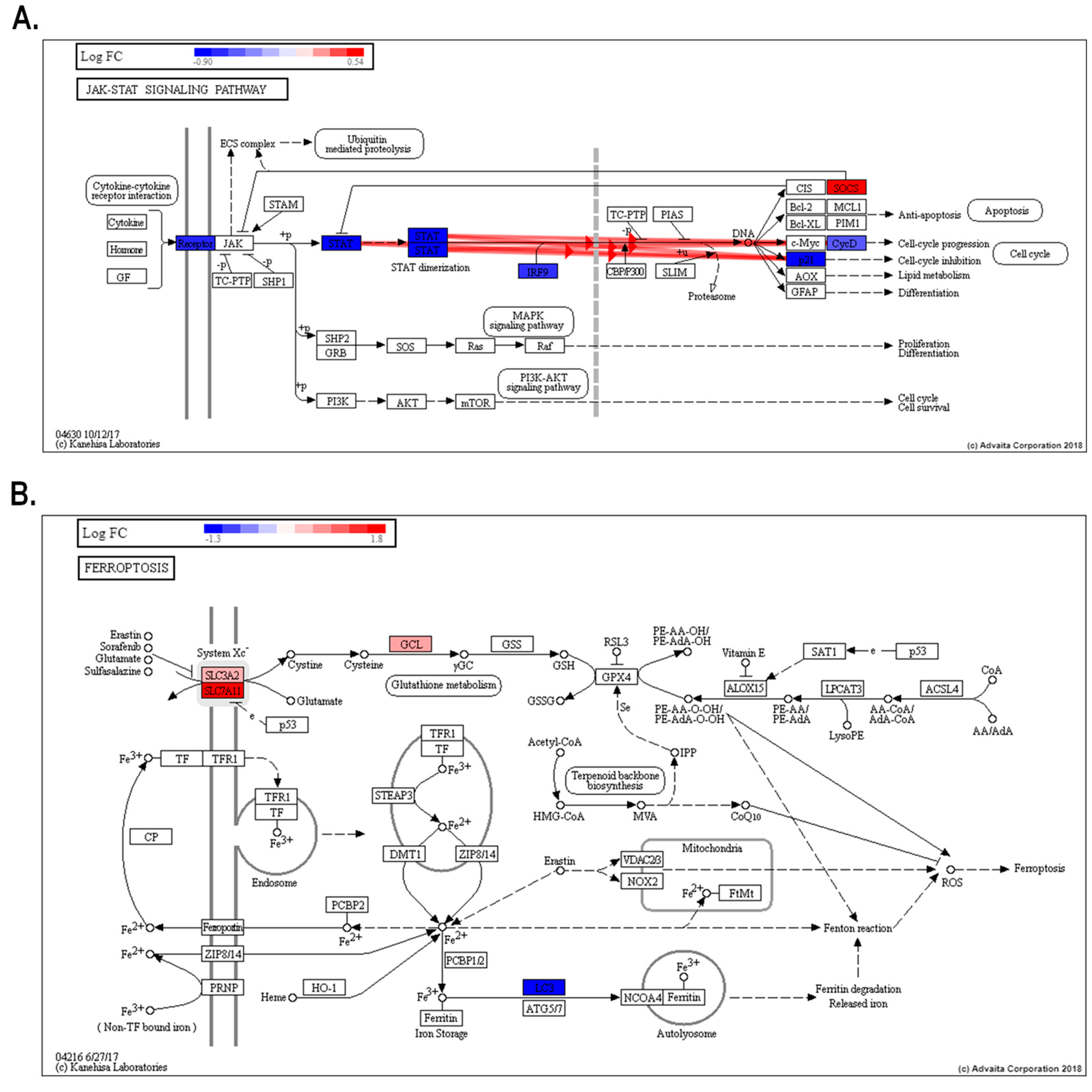
Figure 5.
JAK-STAT and ferroptosis signaling pathways are important for avicequinone B-induced cytotoxicity of CRC cells. Treatment with avicequinone B (8.20 µM) significantly impacts the expression of genes involved in JAK-STAT and ferroptosis signaling pathways, both included in the top twenty-five impacted pathways. (A) The inhibition of JAK-STAT pathway might explain the remarkable suppression of interferon regulated genes (IRGs); (B) while cellular death trough ferroptosis fits with the increased Sytox staining and oxidative stress induced by test compound. Impact analysis was performed using iPathwayGuide online software. The heat map at the top left shows the Log2 fold change (LogFC), where blue indicates down-regulated genes and red indicates up-regulated genes. Only those genes that meet our criteria of differential expression are shown (Log2ǀ(fold change)ǀ≥0.5, P value ≤ 0.001, and q -value ≤ 0.05). The bold red highlighted arrows indicate coherent cascades.
.
JAK-STAT and ferroptosis signaling pathways are important for avicequinone B-induced cytotoxicity of CRC cells. Treatment with avicequinone B (8.20 µM) significantly impacts the expression of genes involved in JAK-STAT and ferroptosis signaling pathways, both included in the top twenty-five impacted pathways. (A) The inhibition of JAK-STAT pathway might explain the remarkable suppression of interferon regulated genes (IRGs); (B) while cellular death trough ferroptosis fits with the increased Sytox staining and oxidative stress induced by test compound. Impact analysis was performed using iPathwayGuide online software. The heat map at the top left shows the Log2 fold change (LogFC), where blue indicates down-regulated genes and red indicates up-regulated genes. Only those genes that meet our criteria of differential expression are shown (Log2ǀ(fold change)ǀ≥0.5, P value ≤ 0.001, and q -value ≤ 0.05). The bold red highlighted arrows indicate coherent cascades.
Collectively, these results suggest that IRGs might play a central regulatory role in the bioactivity of avicequinone B. Over the past decades, the idealization of IFNs, especially INF-γ, as an immunologic guardian against malignant neoplastic disease has changed.17 As a fact, IFNs act directly on cancer cells to inhibit proliferation, modulate apoptosis, improve antigen expression, and up-regulate immune interacting molecules; which is complemented indirectly by activating immune cells, inhibiting immune-suppressive cells, and promoting the switch of tumor associated macrophages (TAMs).18 However, like most cytokines, IFNs induce feedback inhibitory mechanisms to restrain the magnitude of immune responses to avoid the destruction of tissues; therefore, in the scenario where the suppression overcomes the activating mechanisms, IFNs might enhance proliferative and anti-apoptotic signals in tumor cells providing a survival advantage to cancer cells. Indeed, in tumors IFN-γ induces the expression of inhibitory receptors, such as programmed cell death 1 ligand 1 (PDL1) on tumor cells and TAMs or suppressor of cytokine signaling 2 (SOCS2) in dendritic cells.19 In agreement, several recent studies demonstrate the association of treatment-resistant cancer cells and the overexpression of IRGs.
The striking emergence of IFNs and IRGs as important players in the progression and the resistance of cancer cells have gained interest, especially in the context of CRC, since increasing evidence reveals that affected tissue or cells from patients exhibit overexpression of IRGs when compared to normal samples. For example, IRGs such as CMPK2,20,21 IFITM1,22-26 IFITM2,22,27 IFITM3,22,28 IRF2,29 ISG15,30 MX1,31,32 and PLSCR1,33 have been proposed as markers for diagnosis, aggressiveness and prognosis of CRC. Interestingly, the treatment of HT-29 cells with avicequinone B diminished the expression of several CRC-related IRGs (CMPK2, IFIT1, IFITM1, IFITM2, IFITM3, ISG15, and PLSCR1) together with other cancer-related IRGs (IFI27, IFIT2, IFIT3, IRF7, IRF9, and STAT1).
Although in several cases the cellular source of IFNs or IRGs has not been well-defined, intestinal epithelial cells play an important role as producers of inflammatory cytokines (including IFNs) to stimulate anti-viral response among other cellular function,34 and might be speculated that its sustained production could initiate and maintain tumor growth. In agreement, Wagner et al32 recently identified that the elevated expression of inflammatory/IFN-regulated genes (i.e. IFIT1, MX1, IFI44L, among others) is the key factor associated with the intrinsic or acquired resistance of KRAS-mutant epithelial CRC cells to mitogen-activated protein kinase (MEK) inhibitors (MEKi), thus explaining the clinical failure of the MEKi trametinib to treat KRAS or BRAF-CRC, while thriving as BRAF-mutant melanoma therapy. Moreover, their experimental data confirmed that the intrinsic inflammatory environment in the colon, which is intensified by oncogenic KRAS and chemotherapy, results in a tumor microenvironment driven by a persistent inflammatory/IFN transcription program that operates to provide resistance to CRC cells allowing them to survive and proliferate. Hence, the successful treatment of CRC, both in vitro and in vivo , with anti-proliferative compounds (bromodomain inhibitors) involved the suppression of IRGs.
Similarly, the effective cytotoxicity against HT-29 cells exerted by avicequinone B, involved a broad inhibition of IRGs expression, which was also accompanied by a significant impact to the canonical Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling pathway, known to be activated by IFNs and other cytokines (Figure 5A). In normal cells, the activation of JAK-STAT signaling is short and quick, whereas its constitutive activation/overexpression leads to oncogenic transformation, tumor cell invasion, and metastasis.35 In particular, elevated expression/activation of JAK, STAT, IRF9, as well as the inhibition of suppressor of cytokine signaling proteins (SOCS), is reported for various cancer types, including CRC.36-39 In HT-29 cells, the treatment with avicequinone B significantly reduced the expression of various proteins involved in this signaling pathway, from receptor to target genes, while inducing the expression of SOCS. Remarkably, naturally occurring naphthoquinones, structurally related to avicequinone B (i.e. substituted furan-fused naphthoquinones), have been recently reported as STAT3 inhibitors with strong anti-proliferative activity,40 thus supporting our findings. Nonetheless, whether avicequinone B inhibits STAT directly or through a regulator remains to be determined. Overall, this evidence indicates that avicequinone B significantly inhibits the core JAK-STAT-IRGs response, resulting in a distinctive down-regulation of IRGs as an important mechanism for it anti-proliferative effect against HT-29 cells.
In addition, avicequinone B significantly impacted other signaling pathways related to cellular proliferation and survival such as phosphatidylinositol-3’-kinase (PI3K)/serine-threonine kinase (AKT) pathway and the mitogen-activated protein kinase (MAPK) pathway. In this regard, the extracellular signal-regulated kinase (ERK) was identified as an important molecular target suppressed by avicequinone B. Notably, ERK activation/expression is involved in the cellular response to environment regulating cell cycle and cellular differentiation and senescence, which is critical for cancer initiation and progression (Figure S4). In agreement, Patreep et al8 demonstrated that the inhibition of ERK phosphorylation by avicequinone B is an important mechanism involved in the induction of detachment-induced cell death, also known as anoikis, in resistant lung cancer cells. Taken together, this experimental evidence demonstrates that ERK-related signaling pathways play an important role in the cytotoxic effect of the test compound against cancer cells.
Another pathway impacted by avicequinone B was ferroptosis, an iron-dependent non-apoptotic and non-necrotic oxidative form of programmed cell death that involves lethal, iron-catalyzed lipid peroxidation. Ferroptosis can be induced by two classes of small-molecule substances known as class 1, inhibitors of the cystine/glutamate antiporter (system Xc− SLC3A2/SLC7A11) that reduces glutathione (GSH) content, and class 2 or inhibitors of the glutathione peroxidase 4 (GPx4).41 From our results (Figure 5B), it appears that oxidative cell death induced by avicequinone B might be a consequence of (1) a decrease in the Xc− system function (decrease GSH and induces SLC7A11/SLC3A2) or (2) the induction of oxidative stress, through redox cycling, with the consequent GSH depletion and glutamate cysteine ligase (GLC) overexpression; which are both accompanied by (3) increase of intracellular iron as suggested by the down-regulation of LC3A and the up-regulation of ferritin pseudogenes (FTH1P10 and FTH1P20). To note, avicequinone B shifted the expression of SLC7A11, SLC3A and LC3 in the opposite direction to the alterations found in CRC.42-44 Furthermore, avicequinone B also reduced the expression of HSPB1 (also known as HSP-27), a heat shock protein associated to chemotherapy resistance and poor prognosis in CRC.45,46 Interestingly, HSPB1 protects cells from oxidative stress by reducing cellular iron uptake47,48; likewise its suppression sensitizes cancer cells (HeLa, U2OS, and LNCaP) and human xenografts mouse model to erastin-induced ferroptosis.49 In addition to our transcriptomic analysis, the increased influx of SYTOX Green, a membrane-impermeable nucleic acid dye that labels cells with ruptured plasma membranes, is consistent with the induction of ferroptosis, as previously described by Kim et al. 50 Collectively, our results suggest that avicequinone B might induce ferroptosis as a novel mechanism to inhibit cellular proliferation.
Alternatively, the analysis of the gene expression data predicted the presence of 5 miRNAs (hsa-miR-590-5p; hsa-miR-21-5p; hsa-miR-329-3p; hsa-miR-362-3p; and hsa-miR-1306-5p) based on enrichment of differentially down-regulated target genes. Among them, hsa-miR-21-5p arises as a noteworthy molecular target for avicequinone B. Although the prediction of this miRNA might appear as a deleterious side-effect of avicequinone B treatment, since it is a well-characterized oncogenic miRNA in CRC51; it could also indicate that test compound is inducing a severe cellular stress that might stimulate its production as a compensatory mechanism52; or that avicequinone B might be promoting a direct induction of miR-21-5p sensitizing HT-29 cells to death. This is supported by experimental evidence of CRC cells that exhibited higher sensitivity to chemoradiation treatment by overexpression of miR-21-5p.53 Furthermore, berberine (a natural isoquinoline alkaloid) suppressed growth and induced apoptosis in HepG2 cells, through miR-21 overexpression.54
Conclusion
To sum up, our data demonstrated the cytotoxicity of avicequinone B, a natural furan-fused naphthoquinone from Avicennia alba, and its selectivity against several adenocarcinoma cell lines, including HT-29 CRC cells. In this cell line, the anti-proliferative effect of avicequinone B was induced by cell cycle arrest at G2/M and necrosis-like cell death by the induction of an anti-inflammatory gene expression program with special suppression of IRGs probably through blockade of the JAK-STAT-IRGs pathway, combined with the inhibition of proliferative signaling, as well as the induction of ferroptosis and miR-21 expression. Taken together, these results provide novel mechanisms involved in the potential anti-cancer effect of avicequinone B, as well as other substituted fused-naphthoquinones, encouraging further studies to validate its activity using in vivo models.
Ethical Issues
Not applicable.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
This research was funded by Colciencias and the University of Cartagena (Grant 110756933930-2012 to LF and RG). The authors thank the Institute for Immunological Research of the University of Cartagena for their generous assistance with flow cytometry, as well as Carlos Rojas, Julián Nova, Mauro Narváez, and Jesús Cantillo for their help with genotoxicity evaluation.
Supplementary Materials
Supplementary file 1 contains supplementary methods, Tables S1-S5 and Figures S1-S6.
(pdf)
References
-
Kerry RG, Pradhan P, Das G, Gouda S, Swamy MK, Patra JK. Anticancer potential of mangrove plants: neglected plant species of the Marine ecosystem. In: Akhtar MS, Swamy MK, eds. Anticancer plants: properties and application. Singapore: Springer; 2018. p. 303-25. 10.1007/978-981-10-8548-2_13
- Lugo AE. Conserving Latin American and Caribbean mangroves: issues and challenges. Madera y Bosques 2016; 8:5-25. doi: 10.21829/myb.2002.801289 [Crossref] [ Google Scholar]
- Thatoi H, Samantaray D, Das SK. The genus Avicennia, a pioneer group of dominant mangrove plant species with potential medicinal values: a review. Front Life Sci 2016; 9(4):267-91. doi: 10.1080/21553769.2016.1235619 [Crossref] [ Google Scholar]
- Han L, Huang X, Dahse HM, Moellmann U, Fu H, Grabley S. Unusual naphthoquinone derivatives from the twigs of Avicennia marina. J Nat Prod 2007; 70(6):923-7. doi: 10.1021/np060587g [Crossref] [ Google Scholar]
- Assaw S, Mohd Amir MIH, Khaw TT, Bakar K, Mohd Radzi SA, Mazlan NW. Antibacterial and antioxidant activity of naphthofuranquinones from the twigs of tropical mangrove Avicennia officinalis. Nat Prod Res 2019:1-4. doi: 10.1080/14786419.2018.15382200 [Crossref]
- Ito C, Katsuno S, Kondo Y, Tan HT, Furukawa H. Chemical constituents of Avicennia alba Isolation and structural elucidation of new naphthoquinones and their analogues. Chem Pharm Bull (Tokyo) 2000; 48(3):339-43. doi: 10.1248/cpb.48.339 [Crossref] [ Google Scholar]
- Sutton DC, Gillan FT, Susic M. Naphthofuranone phytoalexins from the grey mangrove, Avicennia marina. Phytochemistry 1985; 24(12):2877-9. doi: 10.1016/0031-9422(85)80018-2 [Crossref] [ Google Scholar]
- Prateep A, Sumkhemthong S, Karnsomwan W, De-Eknamkul W, Chamni S, Chanvorachote P. Avicequinone B sensitizes anoikis in human lung cancer cells. J Biomed Sci 2018; 25(1):32. doi: 10.1186/s12929-018-0435-3 [Crossref] [ Google Scholar]
- Tseng CH, Chen YL, Yang SH, Peng SI, Cheng CM, Han CH. Synthesis and antiproliferative evaluation of certain iminonaphtho[2,3-b]furan derivatives. Bioorg Med Chem 2010; 18(14):5172-82. doi: 10.1016/j.bmc.2010.05.062 [Crossref] [ Google Scholar]
- Lin KL, Su JC, Chien CM, Tseng CH, Chen YL, Chang LS. Naphtho[1,2-b]furan-4,5-dione disrupts Janus kinase-2 and induces apoptosis in breast cancer MDA-MB-231 cells. Toxicol In Vitro 2010; 24(4):1158-67. doi: 10.1016/j.tiv.2010.02.019 [Crossref] [ Google Scholar]
- Su JC, Lin KL, Chien CM, Tseng CH, Chen YL, Chang LS. Naphtho[1,2-b]furan-4,5-dione inactivates EGFR and PI3K/Akt signaling pathways in human lung adenocarcinoma A549 cells. Life Sci 2010; 86(5-6):207-13. doi: 10.1016/j.lfs.2009.12.006 [Crossref] [ Google Scholar]
- Jia R, Guo YW, Hou HX. Studies on the chemical constituents from leaves of Avicennia marina. Chin J Nat Med 2004; 2(1):16-9. [ Google Scholar]
- Koyanagi J, Yamamoto K, Nakayama K, Tanaka A. A short-step synthesis of naphtho[2,3-b]furan-4,9-dione. J Heterocycl Chem 1994; 31(5):1303-4. doi: 10.1002/jhet.5570310534 [Crossref] [ Google Scholar]
- Rok Lee Y, So Kim B, Ug Jung Y, Soo Koh W, Soon Cha J, Woo Kim N. Facile synthesis of avicequinone-B natural product. Synth Commun 2002; 32(20):3099-105. doi: 10.1081/SCC-120013719 [Crossref] [ Google Scholar]
- Acuña J, Piermattey J, Caro D, Bannwitz S, Barrios L, López J. Synthesis, anti-proliferative activity evaluation and 3D-QSAR study of naphthoquinone derivatives as potential anti-colorectal cancer agents. Molecules 2018; 23(1). doi: 10.3390/molecules23010186 [Crossref]
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc 2006; 1(5):2315-9. doi: 10.1038/nprot.2006.339 [Crossref] [ Google Scholar]
- Zaidi MR, Merlino G. The two faces of interferon-γ in cancer. Clin Cancer Res 2011; 17(19):6118-24. doi: 10.1158/1078-0432.ccr-11-0482 [Crossref] [ Google Scholar]
- Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 2016; 16(3):131-44. doi: 10.1038/nrc.2016.14 [Crossref] [ Google Scholar]
- Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol 2018; 18(9):545-58. doi: 10.1038/s41577-018-0029-z [Crossref] [ Google Scholar]
- Dai W, Li Y, Mo S, Feng Y, Zhang L, Xu Y. A robust gene signature for the prediction of early relapse in stage I-III colon cancer. Mol Oncol 2018; 12(4):463-75. doi: 10.1002/1878-0261.12175 [Crossref] [ Google Scholar]
- Giarnieri E, Alderisio M, Valli C, Vecchione A, Forte A, Turano R. Overexpression of NDP kinase nm23 associated with ploidy image analysis in colorectal cancer. Anticancer Res 1995; 15(5b):2049-53. [ Google Scholar]
- Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res 2006; 66(4):1949-55. doi: 10.1158/0008-5472.can-05-2731 [Crossref] [ Google Scholar]
- He JD, Luo HL, Li J, Feng WT, Chen LB. Influences of the interferon induced transmembrane protein 1 on the proliferation, invasion, and metastasis of the colorectal cancer SW480 cell lines. Chin Med J (Engl) 2012; 125(3):517-22. [ Google Scholar]
- He J, Li J, Feng W, Chen L, Yang K. Prognostic significance of INF-induced transmembrane protein 1 in colorectal cancer. Int J Clin Exp Pathol 2015; 8(12):16007-13. [ Google Scholar]
- Yu F, Xie D, Ng SS, Lum CT, Cai MY, Cheung WK. IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer Lett 2015; 368(1):135-43. doi: 10.1016/j.canlet.2015.07.034 [Crossref] [ Google Scholar]
- Sari IN, Yang YG, Phi LT, Kim H, Baek MJ, Jeong D. Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget 2016; 7(52):86039-50. doi: 10.18632/oncotarget.13325 [Crossref] [ Google Scholar]
- Xu L, Wang R, Ziegelbauer J, Wu WW, Shen RF, Juhl H. Transcriptome analysis of human colorectal cancer biopsies reveals extensive expression correlations among genes related to cell proliferation, lipid metabolism, immune response and collagen catabolism. Oncotarget 2017; 8(43):74703-19. doi: 10.18632/oncotarget.20345 [Crossref] [ Google Scholar]
- Cui K, Wang H, Zai S, Feng Y. [Expression of IFITM3 in colorectal carcinoma and its clinical significance]. Zhonghua Zhong Liu Za Zhi 2015; 37(5):352-5. [ Google Scholar]
- Mei Z, Wang G, Liang Z, Cui A, Xu A, Liu Y. Prognostic value of IRF-2 expression in colorectal cancer. Oncotarget 2017; 8(24):38969-77. doi: 10.18632/oncotarget.17163 [Crossref] [ Google Scholar]
- Fernández LP, Ramos-Ruiz R, Herranz J, Martín-Hernández R, Vargas T, Mendiola M. The transcriptional and mutational landscapes of lipid metabolism-related genes in colon cancer. Oncotarget 2018; 9(5):5919-30. doi: 10.18632/oncotarget.23592 [Crossref] [ Google Scholar]
- Croner RS, Stürzl M, Rau TT, Metodieva G, Geppert CI, Naschberger E. Quantitative proteome profiling of lymph node-positive vs -negative colorectal carcinomas pinpoints MX1 as a marker for lymph node metastasis. Int J Cancer 2014; 135(12):2878-86. doi: 10.1002/ijc.28929 [Crossref] [ Google Scholar]
- Wagner S, Vlachogiannis G, De Haven Brandon A, Valenti M, Box G, Jenkins L. Suppression of interferon gene expression overcomes resistance to MEK inhibition in KRAS-mutant colorectal cancer. Oncogene 2019; 38(10):1717-33. doi: 10.1038/s41388-018-0554-z [Crossref] [ Google Scholar]
- Kuo YB, Chan CC, Chang CA, Fan CW, Hung RP, Hung YS. Identification of phospholipid scramblase 1 as a biomarker and determination of its prognostic value for colorectal cancer. Mol Med 2011; 17(1-2):41-7. doi: 10.2119/molmed.2010.00115 [Crossref] [ Google Scholar]
- Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol 2011; 186(3):1618-26. doi: 10.4049/jimmunol.1002862 [Crossref] [ Google Scholar]
- Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene 2013; 32(21):2601-13. doi: 10.1038/onc.2012.347 [Crossref] [ Google Scholar]
- Luker KE, Pica CM, Schreiber RD, Piwnica-Worms D. Overexpression of IRF9 confers resistance to antimicrotubule agents in breast cancer cells. Cancer Res 2001; 61(17):6540-7. [ Google Scholar]
- Rigby RJ, Simmons JG, Greenhalgh CJ, Alexander WS, Lund PK. Suppressor of cytokine signaling 3 (SOCS3) limits damage-induced crypt hyper-proliferation and inflammation-associated tumorigenesis in the colon. Oncogene 2007; 26(33):4833-41. doi: 10.1038/sj.onc.1210286 [Crossref] [ Google Scholar]
- Culig Z. Suppressors of cytokine signalling-3 and -1 in human carcinogenesis. Front Biosci (Schol Ed) 2013; 5:277-83. [ Google Scholar]
- Groner B, von Manstein V. Jak Stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol 2017; 451:1-14. doi: 10.1016/j.mce.2017.05.033 [Crossref] [ Google Scholar]
- Löcken H, Clamor C, Müller K. Napabucasin and related heterocycle-fused naphthoquinones as STAT3 inhibitors with antiproliferative activity against cancer cells. J Nat Prod 2018; 81(7):1636-44. doi: 10.1021/acs.jnatprod.8b00247 [Crossref] [ Google Scholar]
- Lu B, Chen XB, Ying MD, He QJ, Cao J, Yang B. The role of ferroptosis in cancer development and treatment response. Front Pharmacol 2017; 8:992. doi: 10.3389/fphar.2017.00992 [Crossref] [ Google Scholar]
- Luque-García JL, Martínez-Torrecuadrada JL, Epifano C, Cañamero M, Babel I, Casal JI. Differential protein expression on the cell surface of colorectal cancer cells associated to tumor metastasis. Proteomics 2010; 10(5):940-52. doi: 10.1002/pmic.200900441 [Crossref] [ Google Scholar]
- Giatromanolaki A, Koukourakis MI, Harris AL, Polychronidis A, Gatter KC, Sivridis E. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J Clin Pathol 2010; 63(10):867-72. doi: 10.1136/jcp.2010.079525 [Crossref] [ Google Scholar]
- Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY, Hung SC. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy 2014; 10(7):1179-92. doi: 10.4161/auto.28679 [Crossref] [ Google Scholar]
- Bauer K, Nitsche U, Slotta-Huspenina J, Drecoll E, von Weyhern CH, Rosenberg R. High HSP27 and HSP70 expression levels are independent adverse prognostic factors in primary resected colon cancer. Cell Oncol (Dordr) 2012; 35(3):197-205. doi: 10.1007/s13402-012-0079-3 [Crossref] [ Google Scholar]
- Tsuruta M, Nishibori H, Hasegawa H, Ishii Y, Endo T, Kubota T. Heat shock protein 27, a novel regulator of 5-fluorouracil resistance in colon cancer. Oncol Rep 2008; 20(5):1165-72. [ Google Scholar]
- Arrigo AP, Virot S, Chaufour S, Firdaus W, Kretz-Remy C, Diaz-Latoud C. Hsp27 consolidates intracellular redox homeostasis by upholding glutathione in its reduced form and by decreasing iron intracellular levels. Antioxid Redox Signal 2005; 7(3-4):414-22. doi: 10.1089/ars.2005.7.414 [Crossref] [ Google Scholar]
- Chen H, Zheng C, Zhang Y, Chang YZ, Qian ZM, Shen X. Heat shock protein 27 downregulates the transferrin receptor 1-mediated iron uptake. Int J Biochem Cell Biol 2006; 38(8):1402-16. doi: 10.1016/j.biocel.2006.02.006 [Crossref] [ Google Scholar]
- Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 2015; 34(45):5617-25. doi: 10.1038/onc.2015.32 [Crossref] [ Google Scholar]
- Kim SE, Zhang L, Ma K, Riegman M, Chen F, Ingold I. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat Nanotechnol 2016; 11(11):977-85. doi: 10.1038/nnano.2016.164 [Crossref] [ Google Scholar]
- Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 2015; 149(5):1204-25 e12. doi: 10.1053/j.gastro.2015.07.011 [Crossref] [ Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med 2009; 13(1):39-53. doi: 10.1111/j.1582-4934.2008.00556.x [Crossref] [ Google Scholar]
- Lopes-Ramos CM, Habr-Gama A, Quevedo Bde S, Felício NM, Bettoni F, Koyama FC. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med Genomics 2014; 7:68. doi: 10.1186/s12920-014-0068-7 [Crossref] [ Google Scholar]
- Lo TF, Tsai WC, Chen ST. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One 2013; 8(9):e75628. doi: 10.1371/journal.pone.0075628 [Crossref] [ Google Scholar]