Advanced pharmaceutical bulletin. 11(1):197-203.
doi: 10.34172/apb.2021.021
Research Article
WT-1, BAALC, and ERG Expressions in Iranian Patients with Acute Myeloid Leukemia Pre- and Post-chemotherapy
Hossein Mehralizadeh 1  , Mohammad Reza Aliparasti 2, Mehdi Talebi 3, Shabnam Salekzamani 4, Shohreh Almasi 2, Morteza Raeisi 5, Mehdi Yousefi 1, AliAkbar Movassaghpour 5, *
, Mohammad Reza Aliparasti 2, Mehdi Talebi 3, Shabnam Salekzamani 4, Shohreh Almasi 2, Morteza Raeisi 5, Mehdi Yousefi 1, AliAkbar Movassaghpour 5, * 
Author information:
1Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
2Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
3Department of Applied Cell Science, School of Advance Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran.
4Department of Nutrition, School of Public Health, Bushehr University of Medical Sciences, Bushehr, Iran.
5Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
*
Corresponding Author: AliAkbar Movassaghpour, Tel: +98 41 33343626, Fax: +98 41 33343844, Email:
movassaghpour@gmail.com
Abstract
Purpose:
Acute myeloid leukemia (AML) is the most prevalent acute leukemia in adults. It possesses different cytogenetic and molecular features. The expression of Wilms tumor-1 (WT1), brain and acute leukemia, cytoplasmic (BAALC) and ETS-related gene (ERG) might be considered as prognostic factors in AML patients. The aim of this study was to determine the mRNA expressions of WT-1, BAALC and ERG genes in bone marrow of mononuclear cells and their effects on complete remission in the Iranian AML patients, pre- and post- chemotherapy.
Methods: Forty AML patients with normal karyotype were evaluated. The mRNA gene expressions were measured with quantitative real-time PCR in bone marrow of mononuclear cells of AML patients at the baseline and after chemotherapy. The subtypes of AML and flow cytometry panel were also assessed. Complete remission (CR) after the treatment was addressed for all patients.
Results: The mRNA expressions of WT-1, BAALC and ERG were significantly decreased after the treatment (p = 0.001, 0.017, 0.036). WT-1 mRNA expression was inversely correlated with CR after chemotherapy (P =0.024). There was also significant correlation between baseline expression of BAALC and CR (P =0.046). No significant correlation was observed between ERG and CR pre- and post- chemotherapy (P =0.464 and 0.781). There was also significant correlation between BAALC mRNA expression and CD34+ (P <0.001).
Conclusion: The present study showed that WT-1 decreased significantly after standard chemotherapy which could have favorable effects on CR. Also, the high expression of BAALC could have a poor prognostic role in AML patients. The identification of these gene expressions can be an efficient approach in targeted therapy among AML patients.
Keywords: Acute myeloid leukemia, WT-1, BAALC, ERG, Gene expression, Chemotherapy
Copyright and License Information
© 2021 The Authors.
This is an Open Access article distributed under the terms of the Creative Commons Attribution (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, as long as the original authors and source are cited. No permission is required from the authors or the publishers.
Introduction
Acute myeloid leukemia (AML) is the most prevalent acute leukemia in adults encompassing 80% of cases of this group.1 It is a malignancy characterized by the abnormal differentiation and proliferation of myeloid stem cells clonal population.2 Some special chromosomal alterations can lead to the formation of chimeric proteins which in turn may increase the likelihood of malignant transformation.3
AML is a heterogeneous disease with possessing different cytogenetic and molecular features which are considered as the key prognostic factors predicting the risk of survival and relapse.3 Recent advances in treatment protocols are based on these prognostic factors contributing to individualized therapy and risk-adapted intensification.4
The Wilms tumor 1 (WT-1) locus in chromosomal region 11p13; encodes a transcription factor highly expressed in the cells of the majority of leukemia patients at diagnosis, and apparently participates in leukemogenesis.5 High expression of WT-1 in acute leukemia has been reported to represent a molecular marker of malignant hematopoiesis and was associated with fewer remissions and poor overall survival.6,7
Another molecular change in AML is the deregulated expression of brain and acute leukemia-cytoplasmic (BAALC) gene. BAALC, which maps on chromosome 8 at 8q22.3, is involved in neuroectodermal and hematopoietic development.8 The expression ofBAALC was reported as a precursor for hematopoiesis like the cluster of differentiation CD34+ in early hematopoietic cells.9 High expression of that was found in bone marrow of AML patients with a normal karyotype10 and identified as anindependent poor prognostic indicator, associated with shorter survival.11
No specific role was explained for BAALC in leukemogenesis, but it was suggested that it blocks myeloid differentiation.12 It was suggested that high expression of BAALC is associated with a poor outcome in AML patients.13 Moreover, ETS-related gene (ERG) located on chromosome band 21q22, is a transcription factor required for normal hematopoiesis.14 The elevated expression of ERG was demonstrated in cytogenetically normal AML patients and has a negative role in the prognosis of AML.15,16 The role of standard treatment in the expression of these genes as well as the effect of post- treatment molecular changes on complete remission (CR) are not clearly investigated in the Iranian population.
In a study in North-East of Iran, the overexpression of BAALC was observed in AML patients aged 2 to 77. The up-regulation of BAALC was reported with survivability reduction.17 In another study, high level of BAALC in pediatric AML patients was associated with lower overall survival rate.18 High expression of WT-1 was also shown in a study on Iranian AML patients.19 However, no study was conducted to evaluate these genes expression in AML at pre- and post-treatment phases.
Therefore, this study was designed to assess the expression of WT-1, BAALC, and ERG in normal karyotype of de novo AML patients and to compare the impact of standard chemotherapy on the expressions of these genes.
Materials and Methods
Patients
The study was performed on 40 normal karyotype de novo AML patients referred to Shahid Ghazi Tabatabai hospital in Tabriz, the Oncology and Hematology center in North-West of Iran, throughout the period between October 2018 and October 2019. The diagnosis of AML was confirmed by morphology and flow cytometry of bone marrow samples. Cytogenetic screening was also performed to select the normal karyotype patients. Patients’ karyotype was analyzed based on bone marrow of leukemic cells metaphases. Furthermore, AML subtypes were addressed based on French–American–British classification. The patients aged > 18 with diagnosis of AML were included in the study. Patients with other types of cancer or with secondary AML were excluded. All patients were undergone standard induction therapy according to 7-3 protocol (7 days with Cytarabine and 3 days with Daunorubicin or idarubicin). The second bone marrow sampling was done 2 weeks after the last chemotherapy. CR was assessed based on blast count less than 5%.
Laboratory analysis
Flow cytometry analysis
Bone marrow samples were collected at diagnosis. The volume of bone marrow samples were 5 to 10 ml and collected on sterile ethylenediaminetetraacetic acid (EDTA) tube. For immunophenotyping analysis BD FACS Calibur (Becton, Dickinson Fluorescence Activated Cell Sorter (Becton, Dickinson Company, CA, USA) was used. The CD markers for Acute Leukemia Panel were: TdT, cy MPO, HLA-DR, GLYCO A, CD2, CD3, CD7, CD10, CD11b, CD13, CD14, CD15, CD19, CD20, CD22, CD33, CD34, CD38, CD41, CD45, CD61, CD117.
RNA extraction and cDNA synthesis
First, mononuclear cells were centrifuged by Ficoll-Hypaque density gradient. Total RNA extraction from mononuclear cells and blasts was done using Trizol Reagent (Invitrogen, USA) according to the manufacturer’s instructions. Total RNA (1 μg) was used for cDNAs synthesis with RevertAid™ H Minus Reverse Transcriptase (RT) (200U), primers (10 μM), dNTPs (1 mM), and RiboLock RNase inhibitor (20 U). The process was conducted at diagnosis and after chemotherapy.
Quantitative real-time PCR
The gene expressions of WT-1, BAALC, and ERG were analyzed by quantitative Real-Time PCR at diagnosis and after chemotherapy by Life Science System (Rotor-Gene 6000). cDNA was diluted by 4-fold, then 2 μL was used in each PCR reaction with the volume of 15μL. It contained 150 nM of primers and 1X FastStart SYBR Green Master (Roche). Sequences of primers are listed in Table 1. The expression levels of β-Actin (ACTB), as a reference gene, were used to calculate relative expression levels. Data were shown as a ratio of the target gene/ACTB. The relative quantification (RQ) was performed by 2-ΔΔCt): expression of target genes / β -actin Treated / Untreated = (1+E) -Ct target gene / (1+E) -Ct β-actin Treated / (1+E) -Ct target gene / (1+E) -Ct β-actin Untreated.
Table 1.
Sequences of primers
|
WT-1
|
Forward |
5’-GATAACCACACAACGCCCATC-3’ |
|
|
Reverse |
5’-CACACGTCGCACATCCTGAAT-3’ |
|
BAALC
|
Forward |
5’-GCCCTCTGACCCAGAAACAG-3’ |
|
|
Reverse |
5’-CTTTTGCAGGCATTCTCTTAGCA-3’ |
|
ERG
|
Forward |
5’-AACGAGCGCAGAGTTATCGT-3’ |
|
|
Reverse |
5’-GTGAGCCTCTGGAAGTCGTC-3’ |
|
β
-Actin
|
Forward |
5’-GCTGTGCTACGTCGCCCTG-3’ |
|
|
Reverse |
5’-GGAGGAGCTGGAAGCAGCC-3’ |
Standard curve
A standard curve was used to determine the efficiency of RT-PCR reactions. To perform the curve, a positive PCR product was diluted serially by 10-fold. The logarithmic of concentrations of customary RT-PCR were plotted against the target gene cycling threshold (Ct) of serial dilution. The efficiencies of WT-1, BAALC, ERG and β-Actin were 96%, 93%, 97% and 96%, respectively.
Statistical analysis
For statistical analyses the SPSS software (ver. 21; IBM Corp., Armonk, NY, USA) was used; the significance of p-values was considered less than 0.05. Normality of data distribution was checked based on descriptive status. Normal-distributed data were expressed as mean ± SD, and non-parametric data were shown as median (75%, 25%). For possible difference in gene expression pre- and post-chemotherapy, Wilcoxon-signed rank test was utilized. The association between gene expression and other variables were performed using Spearman correlation-coefficient test. The differences between non-parametric variables were checked by chi-square test.
Results
Patients
In the present study, 40 de novo AML patients were studied. The mean ± SD age of the patients was 36.75 ± 13.38 years. Fifty percent of patients were male. Patients’ clinical characteristics are shown in Table 2. White blood cell count ranged from 570 to 13.5×103 with the median of 139×103. The minimum and maximum blast count was 153 and 9×10,respectively. The flow cytometry analysis showed that the most and the least patients were in M2 and M3 subtypes with 40% and 10%, respectively. Thirty out of 40 patients were found to be leukemic CD34+ positive.
Table 2.
Basic characteristics of patients
|
Characteristics
|
Median (min, max)
|
| Age |
36.75 ± 13.38* |
| Sex (male) |
20 (50%) |
| WBC count/L |
135×103(570-139×103) |
| Blast count /L |
3525.50 (153.00- 90880.00) |
| Blast (%) |
24.50 (7.00-71.00) |
| Platelet count |
63 × 103(13 ×103 – 575 ×103) |
| RBC |
3.06 (2.27- 4.58) |
| HB |
8.7 (6.8- 11.20) |
| MCV |
92.75 (80.00 – 106.00) |
| Neutrophil |
2.48 ×103(210- 58.9 ×103) |
| Lymphocyte |
4.22 ×103 (200 – 27.4 ×103) |
| FAB classification |
|
| M1 |
6 (15%) |
| M2 |
16 (40%) |
| M3 |
4 (10%) |
| M4 |
8 (20%) |
| M5 |
6 (15%) |
| CD34 |
|
| Positive |
(30)75% |
| Negative |
(10) 25% |
Gene expression
Comparison of WT-1, BAALC and ERG gene expressions in AML patients in pre- and post- chemotherapy phases
The gene expression of WT-1, BAALC and ERG decreased significantly in the post-chemotherapy phase. The median of WT-1, BAALC and ERG pre- and post- chemotherapy is shown in Figure 1.
Correlation between WT-1, BAALC and ERG with CR
There was an inverse significant correlation between post-chemotherapy expression of WT-1 and CR (r= -0.387, P =0.024). Moreover, the expression of BAALC was correlated inversely with CR before chemotherapy (r= -0.318, P = 0.046). There was no significant correlation between ERG and CR pre- and post-chemotherapy (P =0.464 and 0.781) (Table 3).
Table 3.
Correlation between WT-1, BAALC and ERG expressions and complete remission pre- and post-chemotherapy
|
|
Complete Remission
|
|
Pre- chemotherapy
|
Post- chemotherapy
|
|
r
|
P
|
r
|
P
|
|
WT-1
|
0.252 |
0.138 |
-0.387 |
0.024
|
|
BAALC
|
-0.318 |
0.046
|
0.027 |
0.861 |
|
ERG
|
0.118 |
0.464 |
-0.045 |
0.781 |
Gene expressions in subtypes
The expressions of WT-1, BAALC and ERG were categorized in the form of high (>median) and low (≤ median). Table 4 depicts the FAB classification in relation to low and high expression for WT-1, BAALC and ERG. The results showed that, all patients in subtype M3 had high expression of WT-1. Also, in subtype M2, 67% of the patients showed high expression of WT-1. However, there were no significant differences among subtypes of AML in terms of WT-1 expression (P =0.08).
Table 4.
Comparison of WT-1, BAALC and ERG expression among AML subtypes and CD34
|
|
WT-1
expression
|
P
*
|
BAALC
expression
|
P
*
|
ERG
expression
|
P
*
|
|
High expression
|
Low expression
|
High expression
|
Low expression
|
High expression
|
Low expression
|
| FAB classification |
|
|
0.080 |
|
|
0.412 |
|
|
0.411 |
| M1 |
2 (33.3%) |
4 (66.7%) |
|
2 (33.3%) |
4 (66.7%) |
|
4 (66.7%) |
2 (33.3%) |
|
| M2 |
8 (66.7%) |
4 (33.3%) |
|
10 (62.5%) |
6 (37.5%) |
|
10 (62.5%) |
6 (37.5%) |
|
| M3 |
4 (100%) |
0 (0%) |
|
2 (50.0%) |
2 (50.0%) |
|
2 (50.0%) |
2 (50.0%) |
|
| M4 |
2 (25.0) |
6 (75.0%) |
|
2 (25.0%) |
6 (75.0%) |
|
2 (25.0%) |
6 (75.0%) |
|
| M5 |
2 (33.3%) |
4 (66.7%) |
|
4 (66.7%) |
2 (33.3%) |
|
2 (33.3%) |
4 (66.7%) |
|
| CD34 |
|
|
0.457 |
|
|
<0.001 |
|
|
0.028 |
| Positive |
14 (77.8%) |
12 (66.7%) |
|
20 (100%) |
10 (50.0%) |
|
18 (90.0%) |
12 (60.0%) |
|
| Negative |
4 (22.2%) |
6 (33.3%) |
|
0 (0%) |
10 (50%) |
|
2 (10.0%) |
8 (40.0%) |
|
Regarding BAALC, 66.7% of subtype M5 were high expressers. In subtypes M2, the percentages of high and low expressions were similar. For ERG, 66.7% and 62.5% of subtype M1 and M2 were high expressers. There were no significant differences between AML subtypes in terms of ERG and BAALC expressions (P >0.05) (Table 4).
Comparison of WT-1, BAALC and ERG expression in relation to CD34+
Table 4 represents the differences of WT-1, BAALC and ERG expressions in AML patients with CD34 positive (CD34+) in comparison with CD34 negative (CD34-). There were no significant differences in WT-1 expressions between two groups. BAALC and ERG expressions were significantly higher in CD34+ AML patients (P <0.001 and 0.028, respectively).
Correlation between WT-1, BAALC and ERG expression and hematologic characteristics
The results showed that (Table 5),WT-1 expression had inverse and significant correlations with platelet and neutrophil counts (r= -0.349, P =0.037; r= -0.351, P =0.036, respectively). There was also a significant correlation between BAALC expression and blast count at baseline (r=0.337, P =0.034). Furthermore, ERG expression was inversely correlated with platelet count (r= -0.581, P <0.001).
Table 5.
Correlation between WT-1, BAALC and ERG expression and hematologic characteristics
|
|
Platelet
|
WBC
|
Blast count
|
Neutrophil
|
|
r
|
P
|
r
|
P
|
r
|
P
|
r
|
P
|
|
WT-1
|
-0.349 |
0.037 |
0.096 |
0.578 |
0.009 |
0.957 |
-0.351 |
0.036 |
|
BAALC
|
-0.189 |
0.249 |
0.291 |
0.068 |
0.337 |
0.034 |
0.253 |
0.115 |
|
ERG
|
-0.581 |
<0.001 |
0.066 |
0.687 |
0.167 |
0.304 |
-0.170 |
0.295 |
Discussion
In the present study, 35% of the patients reached CR at post-chemotherapy phase. WT-1, BAALC and ERG gene expressions in bone marrow decreased significantly after chemotherapy. The most reduction was observed in WT-1 expression from 1.009 to 0.084. Furthermore, there was an inverse association between the expression of WT-1 post- chemotherapy and CR. Parallel to our study, Ujj et al16 reported that CR was achieved in patients whose WT-1 gene expression changed from positive to negative during therapy. They conclude that the affected level of WT-1 expression after therapy could be a prognostic factor in AML patients. In line with that, previous studies reported that the ratio of WT-1 in pre- and post- chemotherapy phases was considered as a predictor of disease outcome in AML patients who have undergone “3+7” chemotherapy.20 Moreover, in a study by Anderson et al it was shown that a 1 log reduction in WT-1 expression in one month of chemotherapy was significantly correlated with the overall survival; and the level of WT-1 in diagnosis was not found to be a prognostic marker in these patients.4 These results showed that early and deep reduction of WT-1 expression can be used as a useful marker for disease outcome in AML patients. The role of WT-1 in hematologic malignancies has not been clearly understood. Earlier studies have reported the involvement of WT-1 not only in proliferation but also in the inhibition of apoptosis in tumor cell cultures.21 Our findings showed that the standard chemotherapy was partially successful in WT-1 reduction that was correlated with CR. Opposite to WT-1, our results depicted that BAALC expression at diagnosis was inversely correlated with CR. The expression of BAALC decreased significantly after chemotherapy, but the reduction was not as high as the reduction in WT-1. No significant correlation between post- chemotherapy BAALC expression and CR was found. Former studies also found that high BAALC expression at diagnosis had negative prognostic effect in AML patients.22-24, In a study by Soliman, et al9 there was no difference in BAALC expression pre- and post- chemotherapy which was in contrast of ours. However, in line with our findings, high BAALC expression was significantly correlated with lower CR.9 Baldus et al25 suggested that the overexpression of BAALC could determine patients’ resistance to treatment. However, in AML patients who have undergone allogeneic hematopoietic stem cell transplantation, BAALC expression had no prognostic effect.26 BAALC was found to block myeloid differentiation and promote leukemogenesis.12 Furthermore, in pediatric AML patients, BAALC and ERG expressions were shown to associate with low induction remission.27,28 BAALC overexpression was found in a group of genes which were identified as a high risk group for survival with MN1, SPARC, HOPX genes.22
In our study, the levels of ERG decreased after treatment. There was no significant correlation between ERG expression and CR pre- and post- chemotherapy phases. In contrast, high ERG was associated with poor CR in the Soliman study.9 Also, Marcucci et al, in a 5.7 year follow-up study, concluded that ERG expression could be an independent prognostic factor in AML patients.15 In a study of 50 AML patients, CR was different between low and high expression of ERG.29 Moreover, in a meta-analysis of seven studies, it was showed that high ERG expression was associated with lower CR and higher relapse in cytogenetically normal AML patients.14 ERG was involved in cell differentiation, proliferation and apoptosis. It also plays a role in leukemogenesis as a fusion partner with the FUS gene in recurrent AML patients.30,31
Our analysis showed that all patients in subtype M3 had high expression of WT-1 at diagnosis and the least percentage of the patents in WT-1 expression were M1 and M5. Accordingly, in a study of forty-three adult AML patients, the highest and the lowest expressions of WT-1 were found in M3 and M5, respectively.4 However, similar to our study, no significant difference was found in WT-1 expressions among AML subtypes.4 In contrast, in a study in Mashhad, Iran, the highest percentage of WT-1 expression was shown in subtype M1 and the lowest was in M6.19 In our study, there was no patient in M6 subtype.
Highest expression of BAALC and ERG were observed in M5 and M1, respectively. There were also no significant differences among subtypes in terms of BAALC and ERG expressions at diagnosis. In line with our result, in an Egyptian study, the differences in BAALC and ERG expressions among AML subtypes were not significant.9 Also, in this study, the number of patients in M1 was the highest in terms of BAALC expression and in M2 and M3 in terms of ERG expression9 which was different from our study. In a study of Zhang et al,26 the M0 subtype had the highest expressions of BAALC and ERG in AML patients and the lowest BAALC expression was in M5 that was opposite to our study groups.
We also compared the WT-1, BAALC, and ERG expressions in terms of CD34+. The findings showed that only BAALC had significantly higher expression in CD34+ patients which was similar to a study of Damiani et al8 on 175 AML patients in Italy. In another study the expression of BAALC was higher in CD34+ patients with AML.32 However, some studies found no significant difference in CD34+ between high and low expression groups of BAALC.29 There was also no significant difference between CD34+ and CD34- in terms of WT-1 expression in Anderson et al study4 that was similar to our results.
Furthermore, WT-1 expression had significant correlations with platelet and neutrophil counts. Also, BAALC was significantly correlated with blast count. In a study by Zhou et al,33 a substantial amount of bone marrow blast counts were observed in AML patients who highly expressed BAALC compared to low-expressed ones. Also, high expression of BAALC was associated with increased blasts in Chinese AML patients.34 In contrast, a number of former studies found no significant correlation between BAALC expression and blast count.9,20,22
This study had some strengths and limitations. This is the first study that was conducted in North-West of Iran and the patients were selected from the referral hospital to which patients with diverse ethnics referred. However, due to limited financial support, we were unable to evaluate the mutations that are molecular abnormalities in AML patients. Also, the small number of patients was another limitation for our study. Future studies with large sample size and longer duration are warranted.
Conclusion
In conclusion, we found a significant decrease in WT-1, BAALC, and ERG expressions after chemotherapy; the decline in WT-1 expression was significantly correlated with CR. Also, BAALC overexpression at diagnosis could be a prognostic marker in AML patients.
Ethical Issue
The study was approved by Committee of Ethics of Tabriz University of Medical Sciences (IR.TBZMED.REC.1398.897). Written informed consent was obtained from all patients. Written informed consent was obtained from all patients.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
This work has been done as part of the M.Sc. thesis for Hossein Mehralizadeh. This study was supported by Stem Cell Research Center at Tabriz University of Medical Sciences, Iran [NO. 63168]. Authors would like to acknowledge Shahid Ghazi Tabatabai hospital, Stem Cell Research Center, and Department of Immunology at Tabriz University of Medical Sciences (Iran) for their great help and cooperation. We also appreciate all the patients who participated in this study.
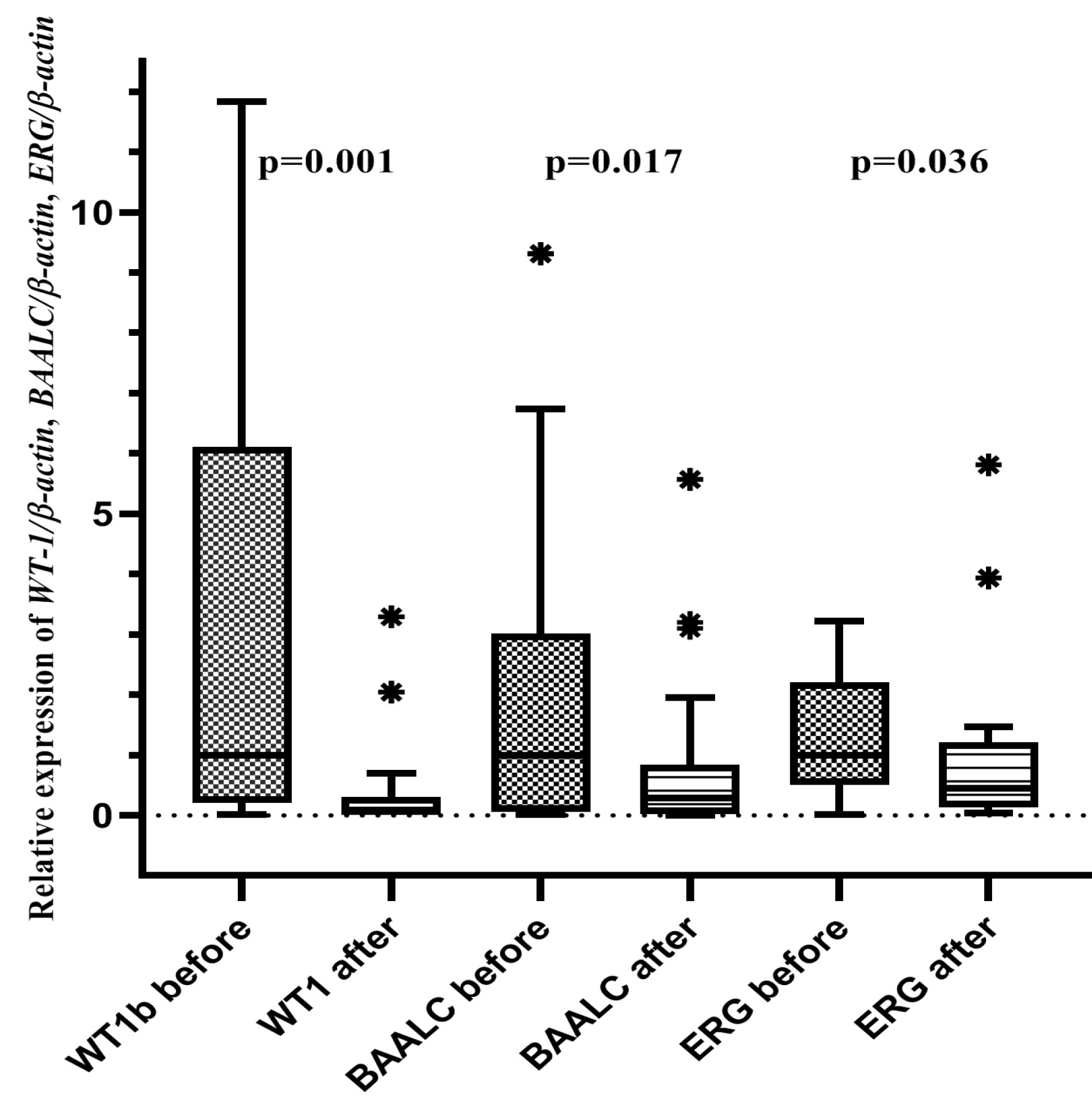
Figure 1.
WT-1, BAALC and ERG expressions in BM of AML patients in pre- and post-chemotherapy phases.
.
WT-1, BAALC and ERG expressions in BM of AML patients in pre- and post-chemotherapy phases.
References
- Rampal R, Figueroa ME. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica 2016; 101(6):672-9. doi: 10.3324/haematol.2015.141796 [Crossref] [ Google Scholar]
- Prada-Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev 2017; 31(1):63-76. doi: 10.1016/j.blre.2016.08.005 [Crossref] [ Google Scholar]
- Mukherjee S, Sekeres MA. Novel therapies in acute myeloid leukemia. Semin Oncol Nurs 2019; 35(6):150955. doi: 10.1016/j.soncn.2019.150955 [Crossref] [ Google Scholar]
- Andersson C, Li X, Lorenz F, Golovleva I, Wahlin A, Li A. Reduction in WT1 gene expression during early treatment predicts the outcome in patients with acute myeloid leukemia. Diagn Mol Pathol 2012; 21(4):225-33. doi: 10.1097/PDM.0b013e318257ddb9 [Crossref] [ Google Scholar]
- Cho BS, Min GJ, Park SS, Shin SH, Yahng SA, Jeon YW. WT1 measurable residual disease assay in patients with acute myeloid leukemia who underwent allogeneic hematopoietic stem cell transplantation: optimal time points, thresholds, and candidates. Biol Blood Marrow Transplant 2019; 25(10):1925-32. doi: 10.1016/j.bbmt.2019.05.033 [Crossref] [ Google Scholar]
-
Li X. Wilms’ Tumor Gene 1 in Different Types of Cancer [dissertation]. Umeå, Sweden: Umeå Universitet; 2015.
- Zhu YM, Wang PP, Huang JY, Chen YS, Chen B, Dai YJ. Gene mutational pattern and expression level in 560 acute myeloid leukemia patients and their clinical relevance. J Transl Med 2017; 15(1):178. doi: 10.1186/s12967-017-1279-4 [Crossref] [ Google Scholar]
- Damiani D, Tiribelli M, Franzoni A, Michelutti A, Fabbro D, Cavallin M. BAALC overexpression retains its negative prognostic role across all cytogenetic risk groups in acute myeloid leukemia patients. Am J Hematol 2013; 88(10):848-52. doi: 10.1002/ajh.23516 [Crossref] [ Google Scholar]
- Soliman A, Abdel Aal A, Afify R, Ibrahim N. BAALC and ERG expression in Egyptian patients with acute myeloid leukemia, relation to survival and response to treatment. Open Access Maced J Med Sci 2016; 4(2):264-70. doi: 10.3889/oamjms.2016.058 [Crossref] [ Google Scholar]
- Bienz M, Ludwig M, Leibundgut EO, Mueller BU, Ratschiller D, Solenthaler M. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res 2005; 11(4):1416-24. doi: 10.1158/1078-0432.ccr-04-1552 [Crossref] [ Google Scholar]
- Yoon JH, Kim HJ, Shin SH, Yahng SA, Lee SE, Cho BS. Implication of higher BAALC expression in combination with other gene mutations in adult cytogenetically normal acute myeloid leukemia. Leuk Lymphoma 2014; 55(1):110-20. doi: 10.3109/10428194.2013.800869 [Crossref] [ Google Scholar]
- Heuser M, Berg T, Kuchenbauer F, Lai CK, Park G, Fung S. Functional role of BAALC in leukemogenesis. Leukemia 2012; 26(3):532-6. doi: 10.1038/leu.2011.228 [Crossref] [ Google Scholar]
- Xiao SJ, Shen JZ, Huang JL, Fu HY. Prognostic significance of the BAALC gene expression in adult patients with acute myeloid leukemia: a meta-analysis. Mol Clin Oncol 2015; 3(4):880-8. doi: 10.3892/mco.2015.562 [Crossref] [ Google Scholar]
- Fang JF, Yuan HN, Song YF, Sun PB, Zheng XL, Wang XJ. E-26 transformation-specific related gene expression and outcomes in cytogenetically normal acute myeloid leukemia: a meta-analysis. Chin Med J (Engl) 2017; 130(12):1481-90. doi: 10.4103/0366-6999.207474 [Crossref] [ Google Scholar]
- Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrózek K, Whitman SP. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol 2005; 23(36):9234-42. doi: 10.1200/jco.2005.03.6137 [Crossref] [ Google Scholar]
- Ujj Z, Buglyó G, Udvardy M, Beyer D, Vargha G, Biró S. WT1 expression in adult acute myeloid leukemia: assessing its presence, magnitude and temporal changes as prognostic factors. Pathol Oncol Res 2016; 22(1):217-21. doi: 10.1007/s12253-015-0002-0 [Crossref] [ Google Scholar]
- Amirpour M, Ayatollahi H, Sheikhi M, Azarkerdar S, Shams SF. Evaluation of BAALC gene expression in normal cytogenetic acute myeloid leukemia patients in north-east of Iran. Med J Islam Repub Iran 2016; 30:418. [ Google Scholar]
- Amirpour M, Ayatollahi H, Sadeghian MH, Sheikhi M, Azarkerdar S, Khiabani A. Analysis of BAALC gene expression as prognosis factor in pediatric acute myeloid leukemia in Iran. Iran J Ped Hematol Oncol 2017; 7(4):237-44. [ Google Scholar]
- Ayatollahi H, Sadeghian MH, Naderi M, Jafarian AH, Shams SF, Motamedirad N. Quantitative assessment of Wilms tumor 1 expression by real-time quantitative polymerase chain reaction in patients with acute myeloblastic leukemia. J Res Med Sci 2017; 22:54. doi: 10.4103/jrms.JRMS_448_16 [Crossref] [ Google Scholar]
- Gianfaldoni G, Mannelli F, Ponziani V, Longo G, Bencini S, Bosi A. Early reduction of WT1 transcripts during induction chemotherapy predicts for longer disease free and overall survival in acute myeloid leukemia. Haematologica 2010; 95(5):833-6. doi: 10.3324/haematol.2009.011908 [Crossref] [ Google Scholar]
- Tuna M, Chavez-Reyes A, Tari AM. HER2/neu increases the expression of Wilms’ Tumor 1 (WT1) protein to stimulate S-phase proliferation and inhibit apoptosis in breast cancer cells. Oncogene 2005; 24(9):1648-52. doi: 10.1038/sj.onc.1208345 [Crossref] [ Google Scholar]
- Torrebadell M, Díaz-Beyá M, Kalko SG, Pratcorona M, Nomdedeu J, Navarro A. A 4-gene expression prognostic signature might guide post-remission therapy in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leuk Lymphoma 2018; 59(10):2394-404. doi: 10.1080/10428194.2017.1422859 [Crossref] [ Google Scholar]
- Weber S, Alpermann T, Dicker F, Jeromin S, Nadarajah N, Eder C. BAALC expression: a suitable marker for prognostic risk stratification and detection of residual disease in cytogenetically normal acute myeloid leukemia. Blood Cancer J 2014; 4(1):e173. doi: 10.1038/bcj.2013.71 [Crossref] [ Google Scholar]
- Yahya RS, Sofan MA, Abdelmasseih HM, Saudy N, Sharaf-Eldein MA. Prognostic implication of BAALC gene expression in adult acute myeloid leukemia. Clin Lab 2013; 59(5-6):621-8. doi: 10.7754/clin.lab.2012.120604 [Crossref] [ Google Scholar]
- Baldus CD, Thiede C, Soucek S, Bloomfield CD, Thiel E, Ehninger G. BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: prognostic implications. J Clin Oncol 2006; 24(5):790-7. doi: 10.1200/jco.2005.01.6253 [Crossref] [ Google Scholar]
- Zhang J, Shi J, Zhang G, Zhang X, Yang X, Yang S. BAALC and ERG expression levels at diagnosis have no prognosis impact on acute myeloid leukemia patients undergoing allogeneic hematopoietic stem cell transplantation. Ann Hematol 2018; 97(8):1391-7. doi: 10.1007/s00277-018-3331-8 [Crossref] [ Google Scholar]
- Aref S, Al Khodary T, Zeed TA, El Sadiek A, El Menshawy N, Al Ashery R. The prognostic relevance of BAALC and ERG expression levels in cytogenetically normal pediatric acute myeloid leukemia. Indian J Hematol Blood Transfus 2015; 31(1):21-8. doi: 10.1007/s12288-014-0395-z [Crossref] [ Google Scholar]
- Hagag AA, El-Lateef AE. Prognostic value of brain and acute leukemia cytoplasmic gene expression in Egyptian children with acute myeloid leukemia. Mediterr J Hematol Infect Dis 2015; 7(1):e2015033. doi: 10.4084/mjhid.2015.033 [Crossref] [ Google Scholar]
- Rashed RA, Kadry DY, El Taweel M, Abd El Wahab N, Abd El Hameed T. Relation of BAALC and ERG gene expression with overall survival in acute myeloid leukemia cases. Asian Pac J Cancer Prev 2015; 16(17):7875-82. doi: 10.7314/apjcp.2015.16.17.7875 [Crossref] [ Google Scholar]
- Oikawa T. ETS transcription factors: possible targets for cancer therapy. Cancer Sci 2004; 95(8):626-33. doi: 10.1111/j.1349-7006.2004.tb03320.x [Crossref] [ Google Scholar]
- Schwind S, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Holland KB. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood 2010; 116(25):5660-9. doi: 10.1182/blood-2010-06-290536 [Crossref] [ Google Scholar]
- Najima Y, Ohashi K, Kawamura M, Onozuka Y, Yamaguchi T, Akiyama H. Molecular monitoring of BAALC expression in patients with CD34-positive acute leukemia. Int J Hematol 2010; 91(4):636-45. doi: 10.1007/s12185-010-0550-8 [Crossref] [ Google Scholar]
- Zhou JD, Yang L, Zhang YY, Yang J, Wen XM, Guo H. Overexpression of BAALC: clinical significance in Chinese de novo acute myeloid leukemia. Med Oncol 2015; 32(1):386. doi: 10.1007/s12032-014-0386-9 [Crossref] [ Google Scholar]
- Qi X, Shen Y, Cen J, Chen H, Sun Y, Sheng H. Up-regulation of BAALC gene may be an important alteration in AML-M2 patients with t( 8;21) translocation. J Cell Mol Med 200 8; 12(6a):2301-4. doi: 10.1111/j.1582-4934.2008.00447.x [Crossref] [ Google Scholar]