Advanced pharmaceutical bulletin. 11(3):557-563.
doi: 10.34172/apb.2021.064
Research Article
Identification of Novel Mutations in Arabidopsis thaliana DOF 4.2 Coding Gene
Omid Jamshidi Kandjani 1  , Mahdieh Rahbar-Shahrouziasl 1, 2, Ali Akbar Alizadeh 3, Maryam Hamzeh-Mivehroud 1, 2, Siavoush Dastmalchi 1, 2, *
, Mahdieh Rahbar-Shahrouziasl 1, 2, Ali Akbar Alizadeh 3, Maryam Hamzeh-Mivehroud 1, 2, Siavoush Dastmalchi 1, 2, * 
Author information:
1Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
2School of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran.
3Pharmaceutical Analysis Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
Abstract
Purpose:
DOF (DNA-binding with One Finger) proteins are plant-specific transcription factors which mediate numerous biological processes. The purpose of the current study is to report new naturally occurring mutations in the gene encoding for one of the members of DOF proteins named DOF 4.2.
Methods:
The expression of zinc finger domain of DOF 4.2 (DOF 4.2-ZF) was investigated by first synthesis of cDNA library using different parts of Arabidopsis thaliana plant. Then the coding sequence for zinc finger domain of DOF 4.2 protein was prepared using nested PCR experiment and cloned into pGEX-6P-1 expression vector. Finally, the prepared construct was used for protein expression. Furthermore, molecular dynamics (MD) simulation was carried out to predict DNA binding affinity of DOF 4.2-ZF using AMBER package.
Results:
For the first time a new variant of DOF 4.2-ZF protein with three mutations was detected. One of the mutations is silent while the other two mutations lead to amino acid replacement (S18G) as well as introduction of a stop codon ultimately resulting in a truncated protein production. In order to investigate whether the truncated form is able to recognize DNA binding motif, MD simulations were carried out and the results showed that the chance of binding of DOF 4.2-ZF protein to cognate DNA in its truncated form is very low.
Conclusion:
The findings demonstrated that the observed mutations adversely affect the DNA binding ability of the truncated form of DOF4.2 if it is expressed in the mutant variant of A. thaliana used in this study.
Keywords: DOF 4.2-ZF, Gene cloning, Protein expression, Homology modeling, Molecular dynamics simulation
Copyright and License Information
©2021 The Authors.
This is an Open Access article distributed under the terms of the Creative Commons Attribution (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, as long as the original authors and source are cited. No permission is required from the authors or the publishers.
Introduction
Different classes of transcription factors have been evolved to specifically contribute to the regulation of plant gene expression as well as mediating plant-specific signals.
1
DOF (DNA-binding with One Finger) proteins are a family of plant-specific transcription factors which contribute to numerous biological processes upon binding to DNA through a highly conserved DNA binding domain. This family of transcription factors has been discovered in both monocots and dicots as well. Genome analysis has revealed that Arabidopsisgenome has 37 putative DOF genes playing essential roles in plant gene expression.
2
DOF proteins are typically composed of a conserved DNA-binding domain and a transcriptional regulation domain organized at N-terminal and C-terminal ends, respectively, which have been linked to each other by conjunctive sequences.
3
The N-terminal DNA-binding domain can recognize various proteins and especially DNA sequences harboring an AAAG core motif.
4,5
Since the discovery of first DOF protein (i.e., ZmDOF1) as a regulator of light response in maize, many attempts have been dedicated to understand DOF proteins versatile roles in biological processes in different plant species.
3
It was later observed that two other maize DOF proteins, namely ZmDOF2 and ZmPBF, contribute in regulation of light response and seed germination, respectively.
6,7
Further studies on DOF family proteins in barely revealed that HvPBF, HvSAD, HvDOF17, and HvDOF19 DOF transcription proteins are involved in regulating the expression of different aleurone hydrolase genes,
8,9
leading to seed germination. In Arabidopsis, DOF proteins act as the mediators of light responses,
10
flowering,
11
plant growth,
12,13
hormone response,
14,15
cell cycle regulation,
16
secondary metabolism,
17
cambium formation and vascular tissue development,
18
as well as seed germination.
15,19
In addition, DOF transcription factors control ammonium assimilation,
20
carbohydrate metabolism,
21
and fatty acid synthesis.
22
DOF transcription factors exert their biological activities through binding either directly or indirectly to the promoters of target genes to regulate downstream signaling pathways. All these facts signify the core regulatory effect of these transcription factors in biological processes determining the amount and quality of plant metabolites and biomass. Several ground-breaking studies have demonstrated that, in contrary to most structural genes, transcription factors are involved in controlling multiple pathways.
23
Therefore, it’s of great importance to explore the function of transcription factors in order to provide understandings applicable for the development of transcription factor-based technology to improve the quality and the efficiency of the plant materials.
23-25
As Arabidopsisgenome sequence has been completed, it is possible to identify putative genes for transcription factors on a genome-wide scale.
2
In the current study we aimed to isolate the gene encoding for DOF 4.2-ZF protein from a native variant of Arabidopsis thalianagrown in greenhouse. However, it was noticed that the used variant carries a truncated form of DOF 4.2-ZF DNA sequence not reported previously. Translation of the mutated DOF 4.2-ZF gene revealed three mutations which were missense, silent and nonsense, resulting in a truncated form of protein. Therefore, in the current study, molecular dynamics simulations were performed to explore the interaction of this truncated form of DOF 4.2-ZF to DNA and compare the results to that of wild type.
Materials and Methods
Cloning of DOF4.2-ZF into pGEX-6P-1 vector
The complete body of
A. thaliana was disrupted for RNA extraction. In order to prevent degradation of RNA, all equipments were double autoclaved to inactivate RNase. QIAGEN RNeasy plant minikit was employed for RNA extraction as described in the manufacturer’s instruction. Briefly, the plant tissue was grounded thoroughly in liquid nitrogen and the obtained powder was decanted into RNase-free liquid-nitrogen–cooled 2 mL microcentrifuge tube. Subsequently, RLT buffer was added to 100 mg of tissue powder and after complete vortexing, the clear lysate was transferred to a QIAshredder spin column and centrifuged for 2 min at 13,000 g. Then, half volume of absolute ethanol was added to the flow-through and the mixture was transferred to an RNeasy Mini spin column and centrifuged for 15 seconds at ≥8000 g. The process was proceeded by addition of Buffer RW1 (700 μL) to the RNeasy spin column followed by centrifugation for 15 seconds at ≥8000 g. The sample was further washed using 500 μL of Buffer RPE. Finally, RNA was eluted from column membrane using 40 μL RNase-free water and kept in -20°C until cDNA synthesis. The cDNA library was synthesized using the prepared mRNA from plant A. thaliana using RevertAid cDNA synthesis kit from Fermentas. For the synthesis of cDNA, 3 μL RNA, 1 μ Loligo-dT and 12 μL DEPC treated water were added into a 1 mL sterile tube and incubated in 70 °C for 5 minutes. Then the tube was placed on ice while 4 μL reaction buffer, 1 μL RNase inhibitor and 2 μL dNTPs were added with additional incubation at 37 °C for 5 minutes. In the next step, 1 μL reverse transcriptase enzyme was added and the mixture was incubated in 42°C for 60 minutes. Finally, the mixture was incubated in 70°C for 10 minutes. Amplification of the desired DNA sequence was carried out using the produced cDNA as the template in the nested PCR reaction as follows: at first, using outer primers, F1 (5’GAAATGAATGTGATGCCCCCACCG 3’) andR1 (5’ATGATTGCCTTGTTGGATCTCAGCC 3’) a larger sequence was amplified and the obtained PCR product was used in the second PCR reaction using inner primers F2 (5’ GAAGGATCCGAAATGAATGTGATGC 3’) and R2 (5’ TTCGGGAATTCTCATTTATCACATATTCC 3’). The product of the latter PCR reaction was electrophoresed on 1% agarose gel followed by gel purification. BamHI and EcoRI restriction enzymes were used to cut the PCR product at 37°C for 4 hours and cloned into the pGEX-6P-1 vector which was already cut with the same enzymes. Using T4 DNA ligase DOF 4.2 ZF domain coding sequence was cloned into pGEX-6P-1 at 16°C for 16 hours and the reaction mixture was transformed into E. coli DH5α strain. The transformed bacteria were cultured on LB-agar plates with 100 µg/mL ampicillin and the obtained single colonies were investigated for the production of DOF 4.2-ZF-pGEX-6P-1 vector. To do so, the single colonies were used to inoculate 10 mL LB medium with 100 µg/mL ampicillin and cultured for overnight at 37°C. In the next day, the bacteria culture was used to extract plasmids using plasmid extraction mini kit. Two PCR reactions were carried out on the extracted plasmid using DOF 4.2-ZF F2 and R2 specific as well as pGEX universal primers to check the validity of ligation process. Finally, the constructed vector was sequenced for final approval.
Expression of DOF4.2 ZF protein
The constructed DOF4.2 ZF-pGEX-6P-1 vector was transformed intoE. coli BL21and a single colony was inoculated into 10 mL LB-ampicillin medium for overnight growth 37°C. The bacterial culture was diluted 1:100 in LB-ampicillin and incubated at 37°C while shaking at 170 rpm. At OD of 0.6, IPTG was added to the culture and the culturing temperature was set to 20°C for overnight protein expression. Different samples before and after induction were taken to monitor protein expression. The next day, the bacteria were separated from medium by centrifugation at 5000 rpm for 15 minutes and suspended in lysis buffer containing Tris 50 mM pH 8, NaCl 150 mM, Triton 1%, lysozyme 0.1 mg/mL, DNAse 10 µg/mL, β-mercaptoethanol 0.1%, PMSF 1.4 mM). Using three cycles of freeze-thaw followed by sonication on ice five times for 30 seconds at 60 pulse the protein content of bacteria was released. In order to separate soluble fraction containing the protein of interest from bacterial debris, the suspension was centrifuged at 10 000 rpm for 30 minutes at 4°C. Expression of protein was evaluated using SDS-PAGE analysis.
Model building and molecular dynamics simulation studies
All sequences and 3D experimental structures were retrieved from UniProt and RCSB databases, respectively. The amino acid sequence for the truncated form of DOF 4.2-ZF protein used in the current study was shown in Figure 1. The 3D model for truncated form of DOF 4.2-ZF protein was derived from the previously proposed model for the wild type protein.
26
The generated model was subjected to molecular dynamics (MD) studies using AMBER suite of programs (version 14) operating on a Linux-based (Centus 6.8) GPU workstation. Initially, the parameter files for the complex of DNA and DOF 4.2 ZF domain were generated using leap module implemented in AMBER program. Then the total charge of the system was neutralized by adding the correct numbers of counter ions (Na+ or Cl−), followed by solvating the system with TIP3P water molecules allowing a buffering distance of 12 Å in all directions in the simulation box. The generated system was equilibrated by applying a brief energy minimization using 500 steps of steepest descent and 500 steps of conjugate gradient minimizations, 50 ps heating step, where the temperature was increased from 0 to 300°K, 50 ps density equilibration, and constant pressure equilibration for 500 ps at 300°K. Using the SHAKE algorithm the bond lengths involving hydrogen atoms were constrained. By employing the particle mesh Ewald (PME) method, the final MD simulation was carried out for 20 ns with a time step of 2 fs. The trajectories were obtained by writing out the coordinates every 10 ps. The RMSD values were calculated using CPPTRAJ module of Amber Tools.

Figure 1.
The DNA and protein sequence alignments between wild type and truncated form of DOF 4.2-ZF. The upper and lower sequence alignments refer to the DNA and protein alignment, respectively. An asterisk (*) indicates positions which have a single, fully conserved residue and a period (.) indicates conservation between groups of weakly similar properties scoring ≤ 0.5 in the Gonnet PAM 250 matrix. The position of mutations is highlighted in gray. In protein sequence alignment, the “X” character indicates end of translation in the truncated DOF-4.2-ZF sequence.
.
The DNA and protein sequence alignments between wild type and truncated form of DOF 4.2-ZF. The upper and lower sequence alignments refer to the DNA and protein alignment, respectively. An asterisk (*) indicates positions which have a single, fully conserved residue and a period (.) indicates conservation between groups of weakly similar properties scoring ≤ 0.5 in the Gonnet PAM 250 matrix. The position of mutations is highlighted in gray. In protein sequence alignment, the “X” character indicates end of translation in the truncated DOF-4.2-ZF sequence.
Results
Cloning of DOF4.2ZF domain sequence into pGEX-6P-1 vector
Twenty-day old
A. thalianaplant was used to extract the total RNA. The extracted RNA was immediately converted to cDNA library, which then used in a nested PCR experiment to amplify the zinc finger domain of DOF4.2 (Figure 2). The product was digested using BamHI and EcoRI restriction enzymes and cloned into pGEX-6P-1 vector (Figure 3). The constructed vector was verified by PCR experiment targeting coding segment for DOF4.2 ZF prior to sequencing (Figure 4). DNA sequencing revealed that the DNA sequence of DOF 4.2-ZF is different in three positions compared to that of wild type A. thalianaspecies (Figure 1). Translation of the DNA sequence to corresponding protein showed that the first mutation is a missense resulting in serine to glycine mutation at position 18 of protein (Figure 1). The second mutation was silent (TAC→TAT) and did not lead to residue type change at position 19(Tyr). In the case of the third mutation, that was a nonsense mutation, the tryptophan codon was changed to stop codon at position 39 in DOF 4.2-ZF sequence. These mutations resulted in a DNA sequence encoding for truncated form of DOF 4.2-ZF (Figure 1).
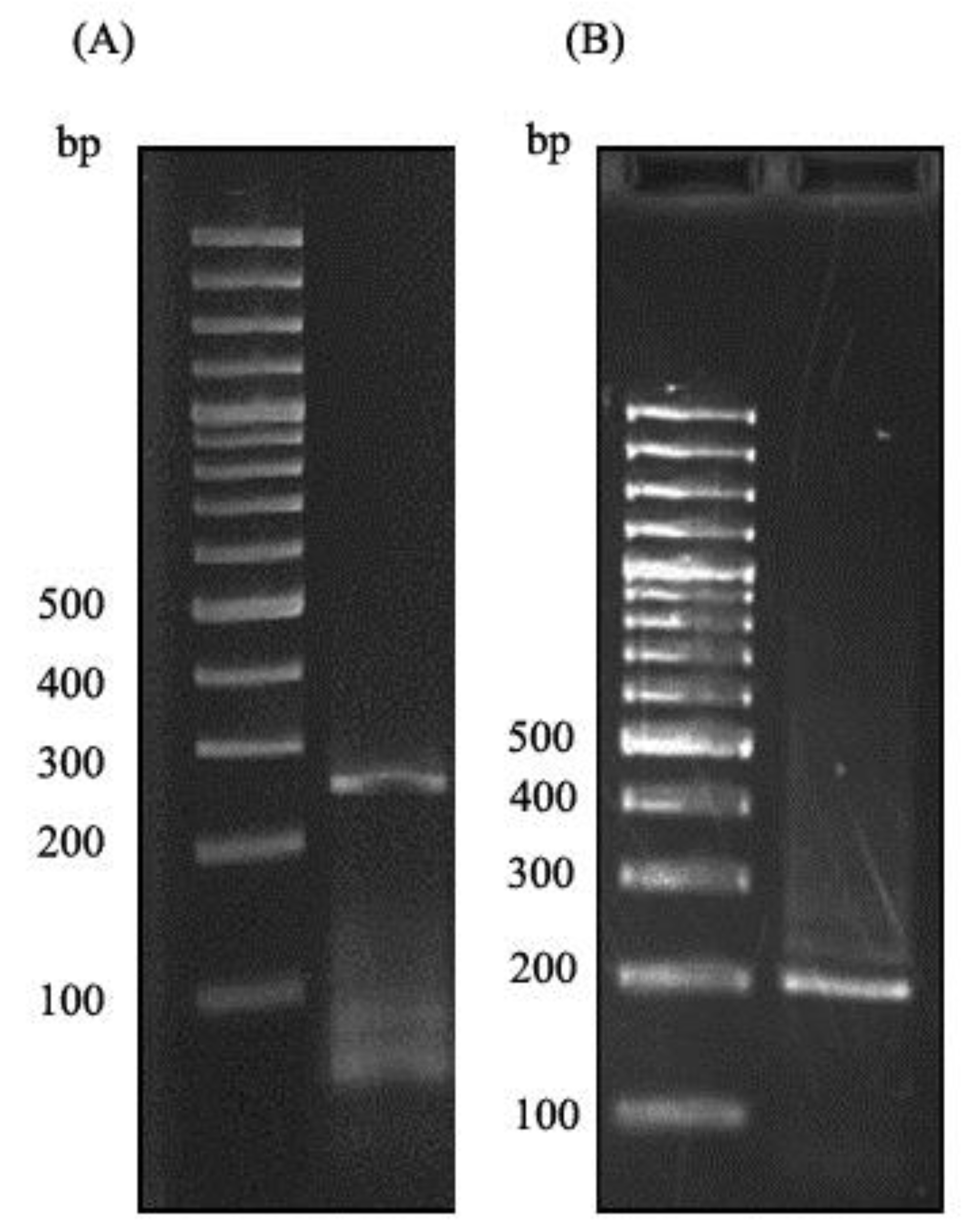
Figure 2.
(A) The gel electrophoresis of PCR product with first set of primers using prepared cDNA library as the DNA template. (B) Gel electrophoresis of second PCR with second set of primers using the PCR product of first PCR as template.
.
(A) The gel electrophoresis of PCR product with first set of primers using prepared cDNA library as the DNA template. (B) Gel electrophoresis of second PCR with second set of primers using the PCR product of first PCR as template.
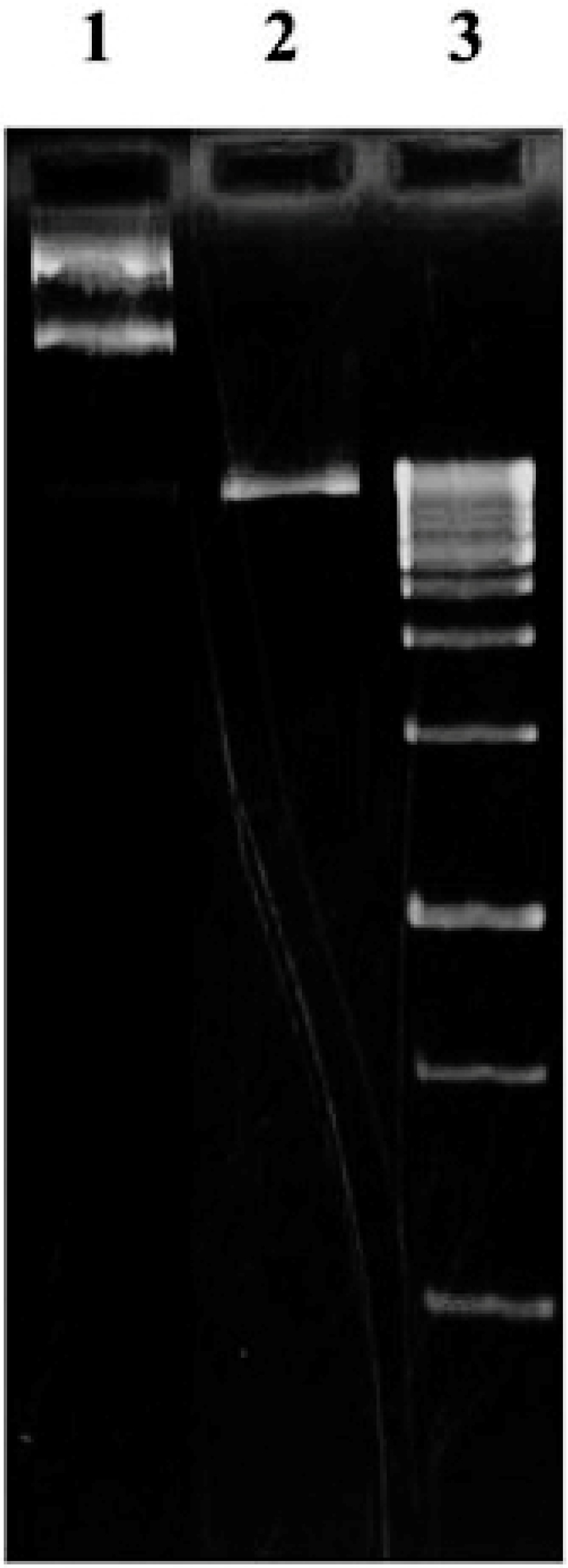
Figure 3.
(1) Undigested pGEX-6P-1 supercoil plasmid, (2) the digested plasmid using BamHI and EcoRI enzymes, (3) DNA ladder.
.
(1) Undigested pGEX-6P-1 supercoil plasmid, (2) the digested plasmid using BamHI and EcoRI enzymes, (3) DNA ladder.
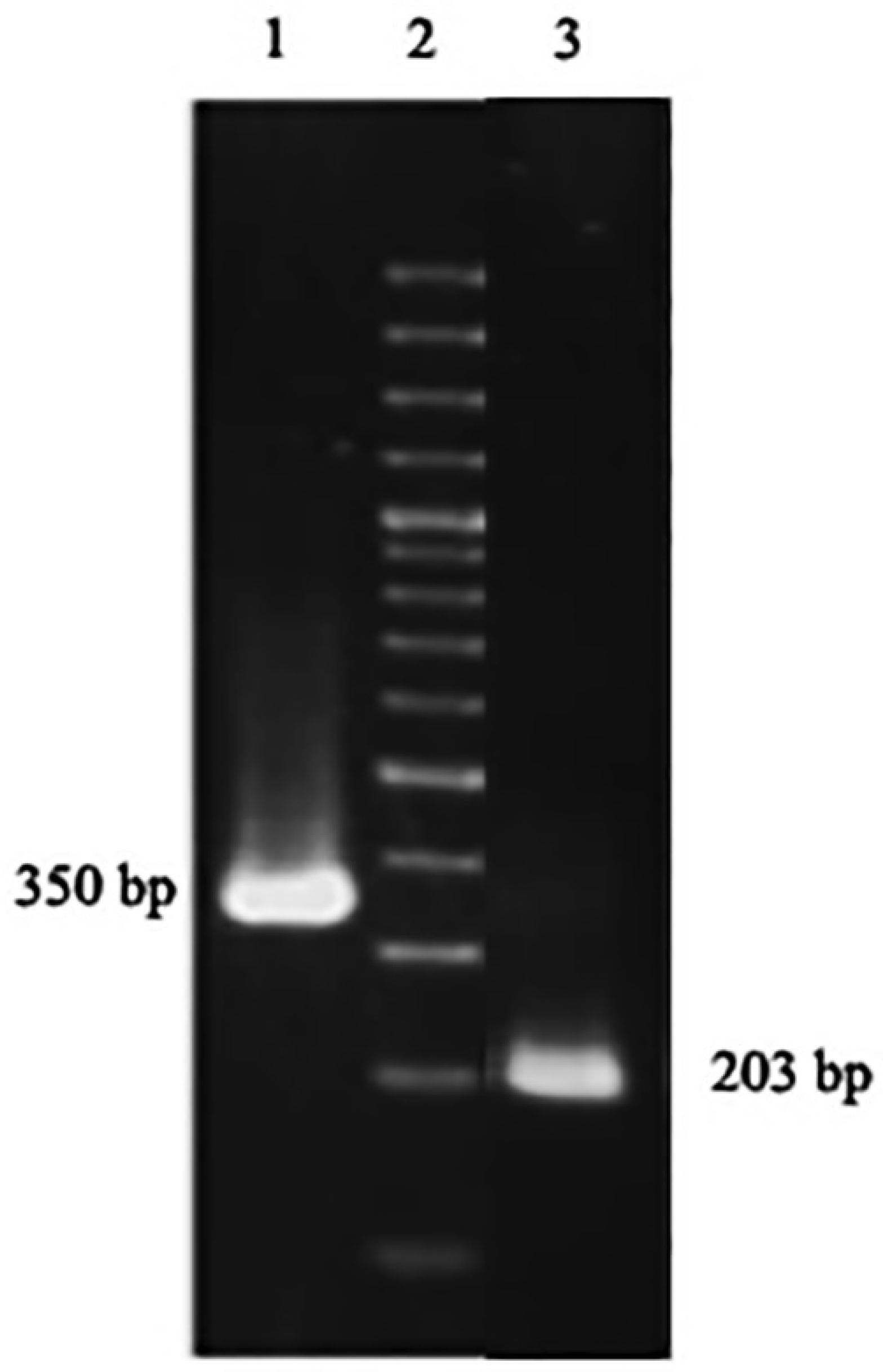
Figure 4.
The gel electrophoresis of PCR products of DOF 4.2-ZF-pGEX-6P-1 construct using universal primers (lane 1), and F2 and R2 set of primers (lane 3).
.
The gel electrophoresis of PCR products of DOF 4.2-ZF-pGEX-6P-1 construct using universal primers (lane 1), and F2 and R2 set of primers (lane 3).
DOF4.2ZF domain expression
The DOF 4.2-ZF domain was expressed in
E. coliBL21 strain. For this purpose, a single colony of E. coli BL21 containing pGEX-DOF 4.2-ZF was used to inoculate 10 mL of LB-ampicillin medium. After overnight incubation, the culture was diluted 1:100 in 100 mL LB-ampicillin and grown to OD of 0.6 before adding IPTG (0.4 mM). The samples taken before and after induction along with the overnight culture were analyzed on SDS-PAGE (Figure 5). The strong band with molecular weight at about 31 kDa stands for truncated DOF4.2-ZF fused to GST tag.
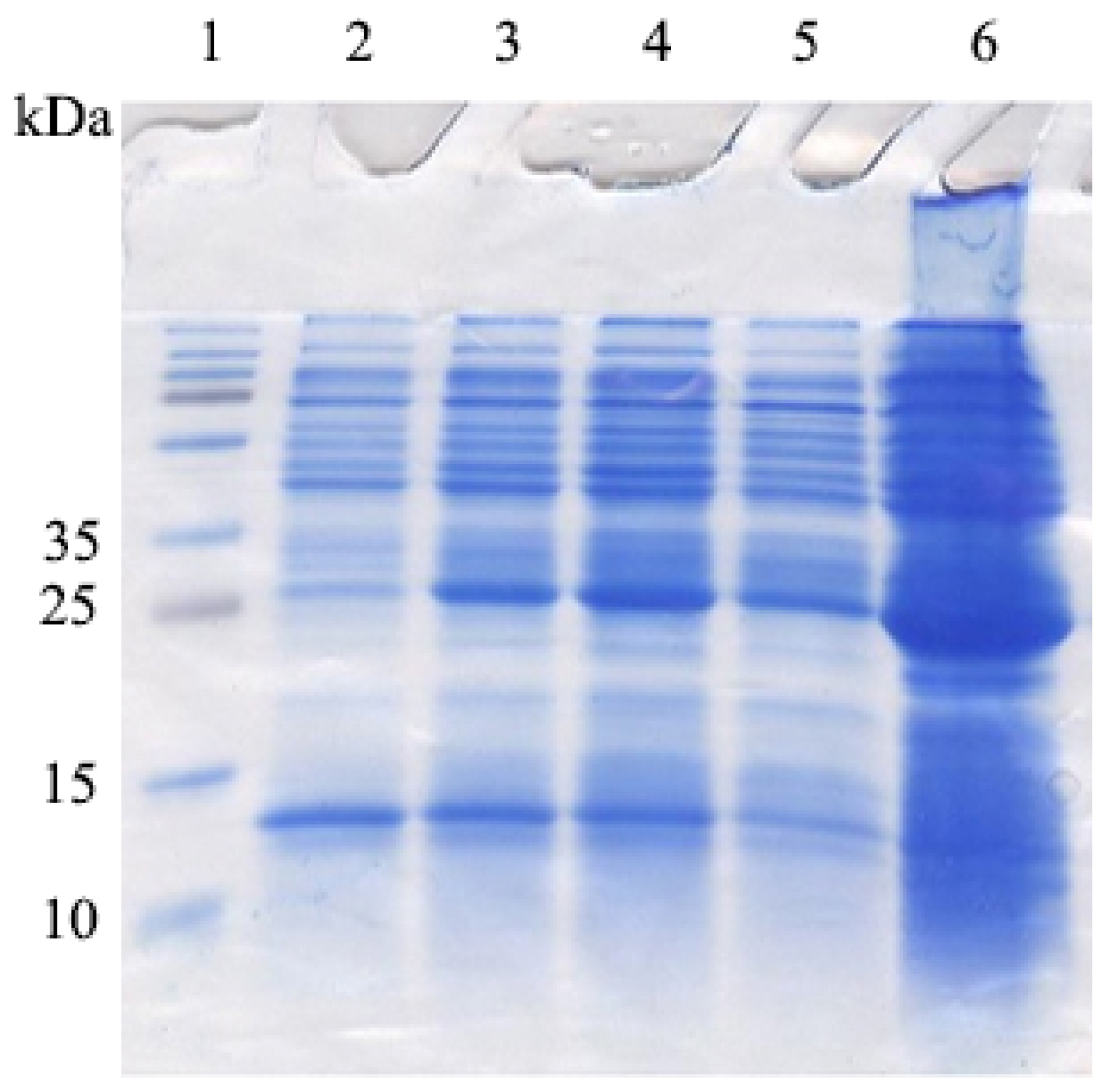
Figure 5.
The SDS-PAGE gel electrophoresis of GST-DOF 4.2-ZF fusion protein. GST-DOF 4.2-ZF fusion protein with the size of 31 kDa can be observed. The lanes are samples prepared from soluble fraction of bacterial lysates: (2) before induction, (3) one hour, (4) two hours, (5) three hours after induction by 0.4 mM IPTG, (6) sample from the overnight culture.
.
The SDS-PAGE gel electrophoresis of GST-DOF 4.2-ZF fusion protein. GST-DOF 4.2-ZF fusion protein with the size of 31 kDa can be observed. The lanes are samples prepared from soluble fraction of bacterial lysates: (2) before induction, (3) one hour, (4) two hours, (5) three hours after induction by 0.4 mM IPTG, (6) sample from the overnight culture.
In silico investigation of DOF 4.2-ZF–DNA interactions
In the current study, it was detected that the in house grown mutant variety of
A. thalianamay express a truncated form of DOF 4.2 protein. Therefore, it was aimed to assess the DNA binding ability of this shortened form using molecular dynamics simulations. To this end, the 3D model of this truncated form was derived from and compared with the previously generated model for the wild type DOF 4.2-ZF built based on GATA1 protein.
26
The stability assessment was carried out using MD simulations for 20 ns in which RSMD values were reached to a plateau after 4 and 13 ns for wild type and truncated DOF 4.2-ZF and remained steady for the rest of the simulations (Figure 6). Additional model validation was performed by analyzing the total energy of the systems which were steady during 20 ns MD simulations revealing the conformational stability of the models (Figure 7). In order to calculate the DNA binding energy for the truncated DOF 4.2-ZF and compare it to that of wild type, MM-GBSA and MM-PBSA algorithms implemented in AMBER suite were used. The results showed that truncated form of DOF 4.2-ZF may not interact with the corresponding DNA deduced from extremely positive value of the calculated binding free energy (Figure 8).
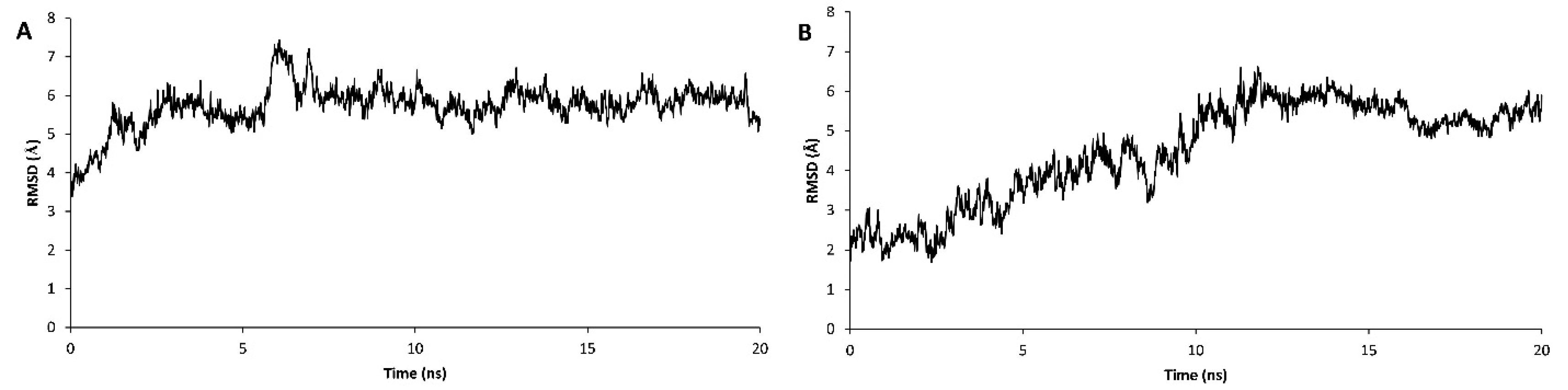
Figure 6.
Plot of root mean square deviation (RMSD) fluctuation for the complex of DOF 4.2-ZF-DNA during 20 ns simulation period. Panels A and B show the results for the wild type and truncated form of DOF 4.2-ZF in complex with DNA, respectively.
.
Plot of root mean square deviation (RMSD) fluctuation for the complex of DOF 4.2-ZF-DNA during 20 ns simulation period. Panels A and B show the results for the wild type and truncated form of DOF 4.2-ZF in complex with DNA, respectively.
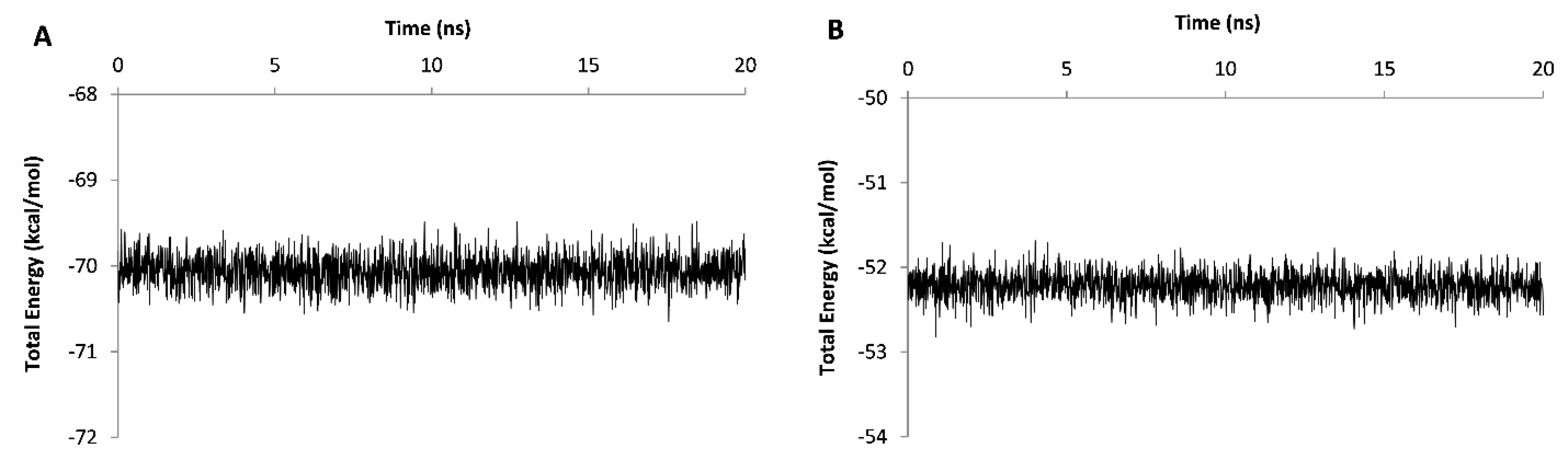
Figure 7.
Plot of potential energy fluctuation for the complex of DOF 4.2-ZF-DNA during 20 ns molecular dynamics simulation period. Panels A and B show the results for the wild type and truncated form of DOF 4.2-ZF in complex with DNA, respectively.
.
Plot of potential energy fluctuation for the complex of DOF 4.2-ZF-DNA during 20 ns molecular dynamics simulation period. Panels A and B show the results for the wild type and truncated form of DOF 4.2-ZF in complex with DNA, respectively.
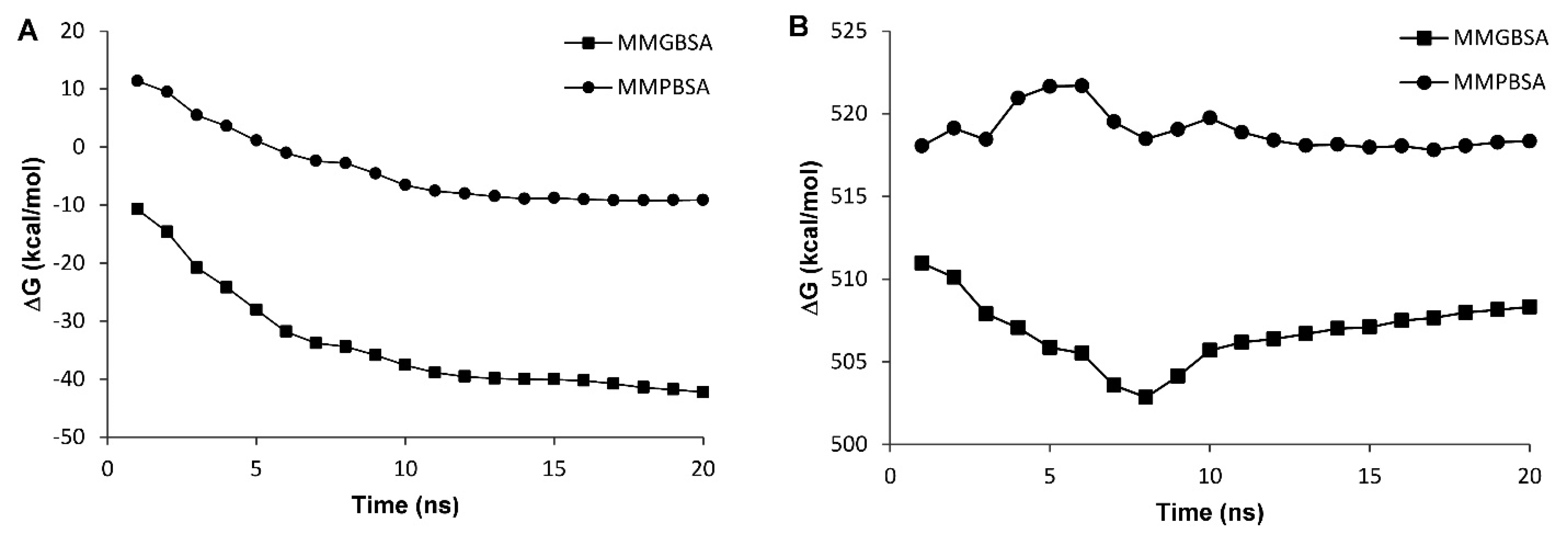
Figure 8.
The plot of binding free energies (∆G) for the DOF 4.2-ZF in complex with DNA during different MD simulation time lengths using MM-PBSA/GBSA calculation methods implemented in AMBER. Panels A and B indicate the results of binding free energies for the wild type and truncated form of DOF 4.2-ZF in complex with DNA, respectively. The filled circles refer to MM-PBSA calculations while the filled squares demonstrate MM-GBSA calculations.
.
The plot of binding free energies (∆G) for the DOF 4.2-ZF in complex with DNA during different MD simulation time lengths using MM-PBSA/GBSA calculation methods implemented in AMBER. Panels A and B indicate the results of binding free energies for the wild type and truncated form of DOF 4.2-ZF in complex with DNA, respectively. The filled circles refer to MM-PBSA calculations while the filled squares demonstrate MM-GBSA calculations.
Discussion
DOF proteins are a family of plant-specific transcription factors which contribute to numerous biological processes such as germination, dormancy, light and defense responses upon binding to DNA through a highly conserved DNA binding domain called DOF domain. In spite of great advances in the field of protein structure determination, efforts to produce and purify DOF domain proteins in order to solve their structures still remains a big challenge as these proteins tend to tightly bind to the corresponding DNA sequences. Taking all these facts into accounts, the current study aimed to produce and purify zinc finger domain of DOF 4.2 for biophysical studies with the aim of identify key interactions involved in DOF-DNA complexes. To do this, total RNA was extracted from A. thalianaplant and used for the synthesis of cDNA library. The gene of zinc finger domain of DOF 4.2 was amplified by a two-step PCR (nested PCR) strategy which can avoid nonspecific amplification of DNA segments related to ZF domains of other DOF proteins (Figure 2). After successful PCR amplification, the gene of interest was cloned at the C-terminus of GST protein using pGEX-6P-1 expression vector. The correctness of the constructed vector was assessed by PCR reactions (Figure 4) and subsequently DNA sequencing. Surprisingly, sequence analysis revealed that there are three nucleotide mutations in the DOF 4.2-ZF DNA sequence compared to the wild type (Figure 1). Translation of the obtained sequence showed that the mutation at position 19 is silent while the two others have resulted in missense mutation at the positions 18 (S18G) and nonsense mutation of tryptophan at position 39 to stop codon (Figure 1). Previous studies have shown the variation of amino acid sequence of DOF 4.2-ZF between the different members of DOF family in two conserved positions including C14S and R32C and the current study demonstrated new mutations in positions 18 and 39 of DOF ZF family, which the latter leads to a truncated form of DOF 4.2 protein. It seems that the aforementioned mutations did not have detrimental effects on germination and growth of the plant.
MD simulations have been extensively used to virtually investigate protein-protein/DNA and protein-small molecules interactions.
27-29
Different studies have used MD simulation based algorithms to report the interaction of DOF family proteins with DNA.
26,30,31
Pandey et al. explored the DNA binding ability of DOF domain from wheat showing that single K29R mutation could adversely affect its binding capability.
31
In another study, we modeled different DOF zinc finger proteins and investigated their ability to bind DNA using MD simulations demonstrating that DOF 3.4 is able to bind DNA with more affinity compared to others.
26
All these evidences show the applicability of MD simulations to calculate the binding energies of DOF protein to DNA in the absence of experimental data. The observation that the variants of A. thaliana used in the current study harbors mutations in the gene for DOF 4.2 protein not reported previously, prompted us to investigate the DNA binding properties of the resulting mutated protein using in silico methods. Analyzing the MD simulation results showed that the binding energies presented as ∆Gbinding values were negative for the wild type indicating a favorable interaction. However, the values for the truncated ZF4.2 were positive (~500 kcal/mol) implying that the truncated form may not bind to DNA spontaneously (Figure 8).
Conclusion
In the current study for the first time we report new variations in gene sequence of DOF 4.2 protein. These variations consist of three mutations in the DNA sequence of DOF 4.2-ZF domain from which one is silent, while the other two are missense and nonsense mutations. The result of these mutations is the production of truncated DOF 4.2 protein, which effectively means the elimination of almost three quarter of residues from its C-terminal. It is noteworthy that the mutant A. thaliana can still grow normally. In the current study, molecular dynamics simulation studies were carried out to compare the DNA binding ability of the produced truncated protein to that of the wild type. The results confirmed that the observed mutations are detrimental for the DNA binding ability of the truncated form of DOF4.2 which might be expressed in the mutant variant of A. thaliana used in this study.
Ethical Issues
Not applicable.
Conflict of Interest
The authors declare that this article content has no conflict of interest.
Acknowledgments
The authors would like to thank the Research Office and Biotechnology Research Center of Tabriz University of Medical Sciences for providing financial support.
References
- Le Hir R, Bellini C. The plant-specific dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis. Front Plant Sci 2013; 4:164. doi: 10.3389/fpls.2013.00164 [Crossref] [ Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2000; 290(5499):2105-10. doi: 10.1126/science.290.5499.2105 [Crossref] [ Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci 2002; 7(12):555-60. doi: 10.1016/s1360-1385(02)02362-2 [Crossref] [ Google Scholar]
- Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem 1993; 268(21):16028-36. [ Google Scholar]
- Zhang B, Chen W, Foley RC, Büttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 1995; 7(12):2241-52. doi: 10.1105/tpc.7.12.2241 [Crossref] [ Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 1998; 10(1):75-89. doi: 10.1105/tpc.10.1.75 [Crossref] [ Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci U S A 1997; 94(14):7685-90. doi: 10.1073/pnas.94.14.7685 [Crossref] [ Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 2005; 42(5):652-62. doi: 10.1111/j.1365-313X.2005.02402.x [Crossref] [ Google Scholar]
- Moreno-Risueno MA, Díaz I, Carrillo L, Fuentes R, Carbonero P. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J 2007; 51(3):352-65. doi: 10.1111/j.1365-313X.2007.03146.x [Crossref] [ Google Scholar]
- Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 2003; 34(2):161-71. doi: 10.1046/j.1365-313x.2003.01710.x [Crossref] [ Google Scholar]
- Wei PC, Tan F, Gao XQ, Zhang XQ, Wang GQ, Xu H. Overexpression of AtDOF47, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol 2010; 153(3):1031-45. doi: 10.1104/pp.110.153247 [Crossref] [ Google Scholar]
- Kang HG, Foley RC, Oñate-Sánchez L, Lin C, Singh KB. Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J 2003; 35(3):362-72. doi: 10.1046/j.1365-313x.2003.01812.x [Crossref] [ Google Scholar]
- Ward JM, Cufr CA, Denzel MA, Neff MM. The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 2005; 17(2):475-85. doi: 10.1105/tpc.104.027722 [Crossref] [ Google Scholar]
- Chen W, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 1996; 10(6):955-66. doi: 10.1046/j.1365-313x.1996.10060955.x [Crossref] [ Google Scholar]
- Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J 2010; 61(2):312-23. doi: 10.1111/j.1365-313X.2009.04055.x [Crossref] [ Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 2008; 56(5):779-92. doi: 10.1111/j.1365-313X.2008.03641.x [Crossref] [ Google Scholar]
- Skirycz A, Jozefczuk S, Stobiecki M, Muth D, Zanor MI, Witt I. Transcription factor AtDOF 4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol 2007; 175(3):425-38. doi: 10.1111/j.1469-8137.2007.02129.x [Crossref] [ Google Scholar]
- Guo Y, Qin G, Gu H, Qu LJ. Dof56/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 2009; 21(11):3518-34. doi: 10.1105/tpc.108.064139 [Crossref] [ Google Scholar]
- Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 2002; 14(6):1253-63. doi: 10.1105/tpc.010491 [Crossref] [ Google Scholar]
- Kumar R, Taware R, Gaur VS, Guru SK, Kumar A. Influence of nitrogen on the expression of TaDof1 transcription factor in wheat and its relationship with photo synthetic and ammonium assimilating efficiency. Mol Biol Rep 2009; 36(8):2209-20. doi: 10.1007/s11033-008-9436-8 [Crossref] [ Google Scholar]
- Tanaka M, Takahata Y, Nakayama H, Nakatani M, Tahara M. Altered carbohydrate metabolism in the storage roots of sweet potato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 2009; 230(4):737-46. doi: 10.1007/s00425-009-0979-2 [Crossref] [ Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 2007; 52(4):716-29. doi: 10.1111/j.1365-313X.2007.03268.x [Crossref] [ Google Scholar]
- Broun P. Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 2004; 7(2):202-9. doi: 10.1016/j.pbi.2004.01.013 [Crossref] [ Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol 2008; 147(1):20-9. doi: 10.1104/pp.108.117887 [Crossref] [ Google Scholar]
- Grotewold E. Transcription factors for predictive plant metabolic engineering: are we there yet?. Curr Opin Biotechnol 2008; 19(2):138-44. doi: 10.1016/j.copbio.2008.02.002 [Crossref] [ Google Scholar]
- Hamzeh-Mivehroud M, Moghaddas-Sani H, Rahbar-Shahrouziasl M, Dastmalchi S. Identifying key interactions stabilizing DOF zinc finger-DNA complexes using in silico approaches. J Theor Biol 2015; 382:150-9. doi: 10.1016/j.jtbi.2015.06.013 [Crossref] [ Google Scholar]
- Alizadeh AA, Jafari B, Dastmalchi S. Alignment independent 3D-QSAR studies and molecular dynamics simulations for the identification of potent and selective S1P1 receptor agonists. J Mol Graph Model 2020; 94:107459. doi: 10.1016/j.jmgm.2019.107459 [Crossref] [ Google Scholar]
- Yang Z, Wu F, Yuan X, Zhang L, Zhang S. Novel binding patterns between ganoderic acids and neuraminidase: insights from docking, molecular dynamics and MM/PBSA studies. J Mol Graph Model 2016; 65:27-34. doi: 10.1016/j.jmgm.2016.02.006 [Crossref] [ Google Scholar]
- Zarei O, Hamzeh-Mivehroud M, Benvenuti S, Ustun-Alkan F, Dastmalchi S. Characterizing the hot spots involved in RON-MSPβ complex formation using in silico alanine scanning mutagenesis and molecular dynamics simulation. Adv Pharm Bull 2017; 7(1):141-50. doi: 10.15171/apb.2017.018 [Crossref] [ Google Scholar]
- Pandey B, Grover A, Sharma P. Molecular dynamics simulations revealed structural differences among WRKY domain-DNA interaction in barley (Hordeum vulgare). BMC Genomics 2018; 19(1):132. doi: 10.1186/s12864-018-4506-3 [Crossref] [ Google Scholar]
- Pandey B, Grover A, Sharma P. Dynamics of Dof domain-DNA interaction in wheat: insights from atomistic simulations and free energy landscape. J Cell Biochem 2018; 119(11):8818-29. doi: 10.1002/jcb.27132 [Crossref] [ Google Scholar]