Advanced pharmaceutical bulletin. 12(1):58-76.
doi: 10.34172/apb.2022.004
Review Article
Isothermal Amplification of Nucleic Acids Coupled with Nanotechnology and Microfluidic Platforms for Detecting Antimicrobial Drug Resistance and Beyond
Seyedeh Zahra Alamolhoda 1, 2  , Nosratollah Zarghami 1, Houman Kahroba 3, Ahmad Mehdipour 4, Mohammad Pourhassan-Moghaddam 1, Rana Jahanban-Esfahlan 1, *
, Nosratollah Zarghami 1, Houman Kahroba 3, Ahmad Mehdipour 4, Mohammad Pourhassan-Moghaddam 1, Rana Jahanban-Esfahlan 1, *  , Morteza Milani 1, 5, *
, Morteza Milani 1, 5, * 
Author information:
1Department of Medical Biotechnology, School of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran.
2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran.
3Department of Molecular Medicine, School of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran.
4Department of Tissue Engineering, School of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran.
5Infectious and Tropical Diseases Research Center, Tabriz University of Medical Science, Tabriz, Iran.
Abstract
Antibiotic resistance is one of the serious health-threatening issues globally, the control of which is indispensable for rapid diagnosis and treatment because of the high prevalence and risks of pathogenicity. Traditional and molecular techniques are relatively expensive, complex, and non-portable, requiring facilities, trained personnel, and high-tech laboratories. Widespread and timely-detection is vital to the better crisis management of rapidly spreading infective diseases, especially in low-tech regions and resource-limited settings. Hence, the need for inexpensive, fast, simple, mobile, and accessible point-of-care (POC) diagnostics is highly demanding. Among different biosensing methods, the isothermal amplification of nucleic acids is favorite due to their simplicity, high sensitivity/specificity, rapidity, and portability, all because they require a constant temperature to work. Isothermal amplification methods are utilized for detecting various targets, including DNA, RNA, cells, proteins, small molecules, ions, and viruses. In this paper, we discuss various platforms, applications, and potentials of isothermal amplification techniques for biosensing of antimicrobial resistance. We also evaluate the potential of these methods, coupled with the novel and rapidly-evolving platforms offered by nanotechnology and microfluidic devices.
Keywords: Biosensing, Isothermal amplification techniques, Antibiotic drug resistance, Nanotechnology, Microfluidics
Copyright and License Information
©2022 The Authors.
This is an Open Access article distributed under the terms of the Creative Commons Attribution (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, as long as the original authors and source are cited. No permission is required from the authors or the publishers.
Introduction
Significance of antimicrobial resistance
Antimicrobial resistance (AMR), as a highly concerning issue all over the world, affected the health programs in countries. The significance of AMR makes the United States, National Institute of Health,
1
World Health Organization (WHO),
2
Centers for Disease Control and Prevention (CDC), and the United Nations (UN) to deal with this issue. The increasing rate of AMR and the emergence of new pathogens from bacteria to viruses, as the coronavirus disease 2019 (COVID-19) pandemic, is a major challenge.
3-5
Extensive growth in antibiotic resistance incidence is confirmed by WHO’s new Global Antimicrobial Surveillance System (GLASS) studies by evaluating 500 000 suspected cases with bacterial infection across 22 different countries. Antibiotic resistance to at least one of the common antibacterial agents was apparent among suspected cases for sepsis ranging from 0-82%. Resistance to penicillin as the common antibacterial agent for pneumonia treatment ranged from 0%-51%. Also, 8%-65% of all diagnosed Escherichia coliinfections were resistant to ciprofloxacin as the standard medicine in treating urinary infection.
6
Not to forget the development of resistance to Mycobacterium tuberculosis, the causative agent for tuberculosis (TB), followed by WHO since 1994. Not to mention that TB as a dangerous and life-threatening disease, affects about one-third of the world’s population.
7
CDC Threats Report for AMR in 2019, indicatesthat unsuccessfully-treated bacterial infections due to AMR, which claims at least 700 000 lives per year around the world, is anticipated to reach more than 10 million deaths per year up to 2050, with a burden of US $100 trillion to the global economy by loss of productivity. Only in the USA, more than 2.8 million multidrug-resistant bacterial infections are reported per year, which are responsible for about 35 000 deaths and $20 billion health-care expenditures.
8
Unlike many dedicated efforts to prevent and control infection in the US to reduce the mortality rate, the number of people facing antibiotic resistance is still too high. Moreover, in 2020, WHO ranked AMR as one of the top 10 global public health threats facing humanity, requiring urgent multi-sectoral action to achieve the Sustainable Development Goals (SDGs). AMR’s economic cost is significant, along with death and disability, prolonged hospitalization, and demands on high-cost medicines; also, the financial challenges for those impacted patients is a critical point. Low- or unavailability of effective antimicrobial strategies and agents limit modern medicine’s success rate in treating infections mainly during major surgeries and cancer chemotherapy.
9-12
The emergence and spread of new forms of resistance remain a concern. What’s even more alarming is the con-infection of the unresolved, worldwide infections such as TB with other emerging infections, such as COVID-19. Reports from five different countries indicated that about 6.9% of the patients with COVID-19 have bacterial infections (3.5% diagnosed concurrently and 14.3% post-COVID-19); also this percentage is higher in patients who require intensive critical care. Nevertheless, a US multicenter study revealed that about 72% of the COVID-19 patients are administered antibiotics even with no clinical symptoms, promoting the AMR in bacteria. AMR can be deteriorated under the influence of COVID-19 infection due to the overprescription of antibiotics, continuous misapplication in agriculture, and the dearth of antimicrobials in the development pipeline. Competing global priorities are decreasing the AMR eradication activities, such as measures for multidrug-resistant tuberculosis.
13,14
Polymerase chain reaction (PCR) techniques for detection of AMR
The introduction of PCR was a big evolution for researchers and clinicians in biological and clinical research, which opened a bride gate to a new molecular and biological research and diagnostics world.
15
Regarding its potential for detecting AMR, using a rapid high-throughput PCR, one study reported an evaluation of more than 7500 highly antibiotic-resistant clinical isolates of Pseudomonas aeruginosa, E. coli, Proteus mirabilis,and Klebsiella pneumonia for antibiotic-resistance genes. These isolates were obtained from different hospitals including Europe, Asia, South America, North America, Africa, and Oceania. Authors compared genetic data with phenotypic resistance across cephalosporins, penicillins, aminoglycosides, carbapenems, fluoroquinolones, trimethoprim-sulfamethoxazole, and macrolides. Using PCR, average positive predictive values for genotypic prediction of phenotypic resistance were 92% for P. aeruginosa, 87% for P. mirabilis, 93% for K. pneumonia, 91% for E. coli.
16
Early identification of genes associated with a specific antibiotic resistance mechanism using a PCR-based approach can reduce unnecessary antibiotic consumption by predicting a gain of bacteria resistance within hours.
17
PCR also can impact hospital-based transmission of antibiotic-resistant organisms by allowing for timely implementation of appropriate precautions when an organism is predicted to have a resistance gene.
18
Equally, reverse transcriptase-PCR (RT-PCR) is the number one diagnostic test for detecting COVID-19 across the world.
19-22
Although PCR technology may impact patient care, further studies are required to assess its utility in the patient care setting. The cost-benefit of PCR as the premier method for AMR detection needs to be reevaluated in the account of the facility needs, resources, costs, and the prevalence of resistant isolates, and the need for adequate and trained personal,
23
especially in the resource-limited settings/regions.
3,24
Isothermal amplification methods for the detection of AMR
Since the early 1990s, isothermal nucleic acid amplification tests have emerged as an essential diagnostic tool, with applications in clinical diagnosis, environmental monitoring, and testing food quality.
25
These methods included loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA), helicase-dependent amplification (HDA), strand displacement amplification (SDA), rolling circle amplification (RCA), recombinase polymerase amplification (RPA), and transcription-mediated amplification (TMA).
26
Amplification of nucleic acids using a constant temperature (isothermal) eliminated the need for different temperature cycles, favored enzyme activity, reduced sample consumption, and saved time to produce fast results. Equally, the fully closed and safe micro-structured device can reduce contamination risk.
27
Together, isothermal amplification-based tests are user-friendly, simple, rapid, sensitive, portable, and require less energy than PCR/RT-PCR and can address the need for a simple, quick, available, and culture-free molecular diagnostic method for widespread assessing of AMR.
28,29
To further simplify these methods, some of them analyze the sample’s raw lysates without the need for nucleic acid purification.
30
Besides early pathogen screening and antibacterial resistance identification, isothermal amplification methods are also applied for cancer cell detection. Recent studies are focused on cancer cell detection using isothermal amplification methods, particularly via identifying microRNAs involved in cancer progression, invasion, and metastasis.
31,32
Further, integration of isothermal amplification methods with newly developed microfluidics and nanotechnology-based approaches generated novel tools for the identification of cancer cells and AMR.
33-35
This review aims to discuss the use of different isothermal-based systems for nucleic acid amplification and highlight their potential in the context of detecting AMR. We also discuss the integration of isothermal-based systems with nanotechnology and microfluidics and evaluate their potential to realize miniature yet sensitive biosensing platforms for the screening of antibiotic drug resistance.
Isothermal amplification platforms for detecting AMR
In this section, we review the variety and potential of isothermal-based-biosensing platforms to detect human-related microbial pathogens. As the sensitivity and specificity of these methods vary among different types of the samples and analytical method used, these are discussed within the text.
Recombinase polymerase amplification
RPA is an isothermal method, which denatures the genomic target DNA using recombinase-primer complexes followed by stabilizing the single-stranded DNA (ssDNA) with the help of ssDNA binding proteins (Figure 1). The RPA detection is very similar to Taq-Man hydrolysis probes except that the probe includes tetrahydrofuran, a basic site analog and cleaved by endonuclease IV. The strand displacing Bsu compared with Taq is more resistant to chemical inhibition.
36
Since the proteins are active elements in DNA denaturation, there is no need for high denaturation temperatures. Thus, the reaction occurs in temperatures ranging from 37-42°C and more rapid than the PCR method, usually 5-7 min.
36-38
The excessive sensitivity of RPA allows the technique to detect tens of the target’s copies.
38-41
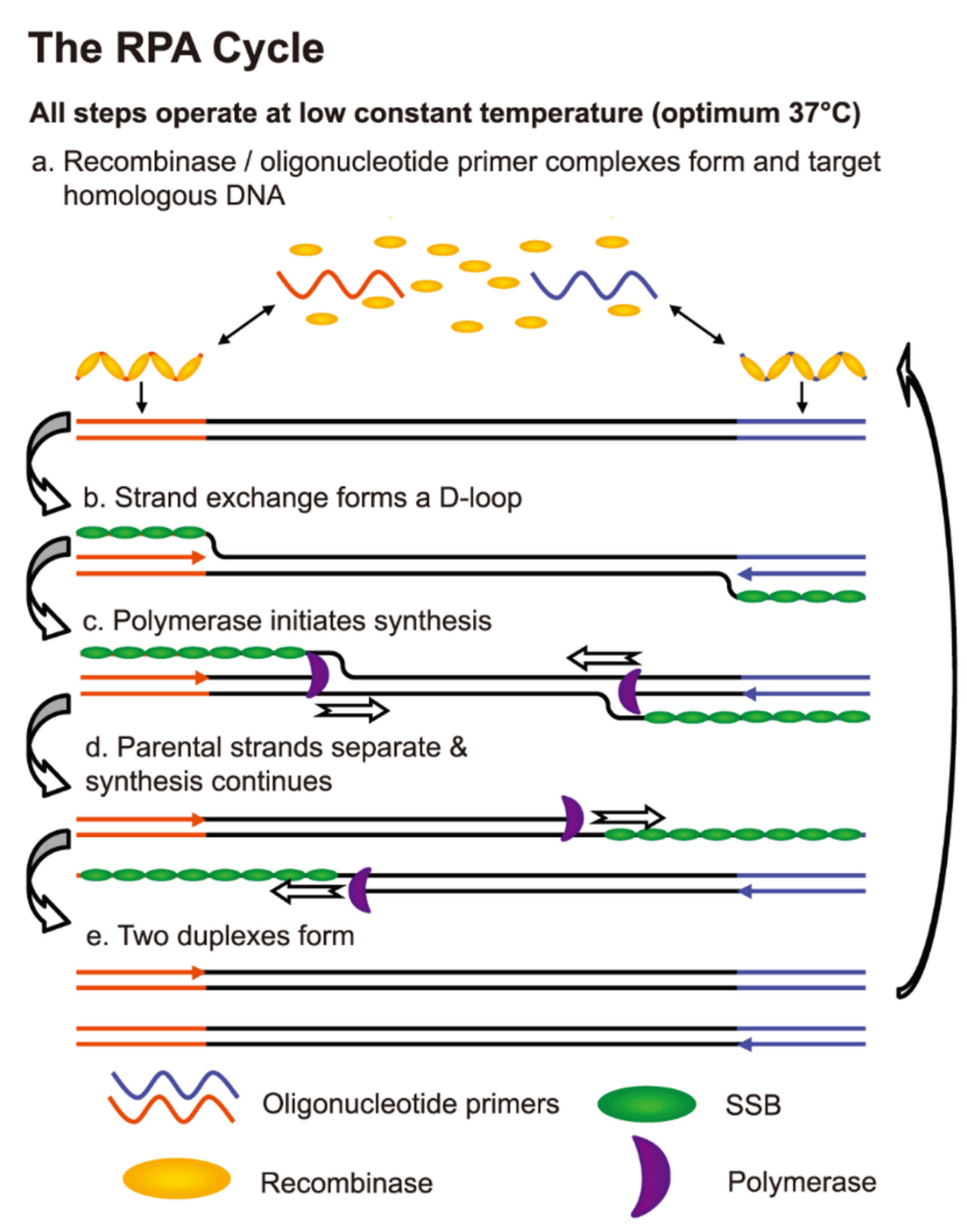
Figure 1.
An overview of RPA for rapid biosensing of TB. SSB: Single-strand binding protein. Adopted from Boyle et al.
42
.
An overview of RPA for rapid biosensing of TB. SSB: Single-strand binding protein. Adopted from Boyle et al.
42
Despite little use of RPA in clinical applications, there is evidence that this method can potentially detect bacterial, viral, and protozoan human pathogens. Moreover, RPA is capable of detecting blood fluke Schistosoma japonicum,
38
Giardia, Cryptosporidium and Entamoeba,
41,43
as well as the human immunodeficiency virus (HIV),
44,45
Chikungunya virus (CHIKV),
37
Rift Valley fever virus,
46,47
foot-and-mouth disease virus (FMDV),
48
Middle East respiratory coronavirus,
49
Crimean Congo hemorrhagic fever virus (CCHFV),
50
and bovine coronavirus.
51
The bacterial species detected by RPA include M. tuberculosis,
52,53
Neisseria gonorrhoeae, methicillin-resistant Staphylococcus aureus(MRSA) and Salmonella enterica.
54
Pectobacterium species are important contributors to soft rot in fruit and vegetable products such as tomatoes and potatoes.
55
So one needs precise and simple methods for rapidly identifying the pathogen for timely management of injuries. The RPA, combined with a lateral flow device (LFD) was developed to detect Pectobacteriumspp. directly from the contaminated plant material without the need for DNA isolation.
56
The specificity of RPA-LFD was determined with 26 Pectobacteriumspp. and 12 non-Pectobacterium species with no false positive or false negative outcomes. The sensitivity and accuracy of the test were determined by the development of RPA primers and probes. Both sensitivity and spiked sensitivity methods obtained the detection limit of 10 fg. When targets were clearly identified from infected potatoes and tomatoes, no inhibitory effect was observed on the RPA assay. Furthermore, this method can also be used to detect reservoir hosts for Pectobacterium species.
57
Moreover, RPA-LFD is used for detection of the plant pathogen Phytophthora sojae, with extraordinary specificity (negative results against 24 other Phytophthora, 14 fungal species, and one Globisporangium), high sensitivity (10 pg/50-μL genomic DNA of P. sojae) within 5 min.
58
Likewise, the RPA-LFD principle is applied for screening of African swine fever virus with the sensitivity of 150 copies per reaction within 10 min at 38°C, and positive rates (10/145) comparable to RT-PCR.
59
RPA is recognized as a particular method in diagnostic and clinical use and resistant to false positives (type I errors). In many cases, a specificity of 100% is indicated.
37,38,40,45
Due to the health risks of diagnostic and therapeutic errors, high specificity is an important feature in diagnostic analyzes. Type II errors (false negatives) may always occur if the pathogenic agent in the sample is low, but accurate RPA sensitivity reduces this error.
60
In one study, Nelson et al
61
developed a new test for RPA to identify the Macrolide Efflux A or mef (A) gene that encodes a pump responsible for causing bacterial resistance to 14- and 15-membered macrolide antibiotics (including erythromycin and azithromycin).
62,63
This gene is also expressed by S. pyogenes, which is located on a transposon that is integrated into a pro-phage.
64,65
This gene was initially identified in S. pyogenes and S. pneumoniae and has so far been identified on a range of Gram-positive and Gram-negative bacteria.
66
Multiple displacement amplification
Multiple displacement amplification (MDA) is another PCR-independent technique to amplify the amounts of DNA with a lower frequency of errors.
67
The test starts by annealing the random hexamer primers to the template DNA followed by a high-fidelity DNA polymerase that works to continue the polymerization and amplification of the target DNA. Phi29 is the most used polymerase enzyme in this method. Since Phi29 is a high processivity enzyme, the MDA method can produce larger-sized products than PCR (greater than 70 kb),
68
rendering it a superior candidate for WGS compared to WGS-PCR, which generates less DNA than 1000 bp long.
69
Moreover, 3’ to 5́ proofreading activity of Phi29 enzyme decreases the error rate to 1 in 106 to 107 basis.
70
The optimum temperature for performing the reaction is 30°C.
71
(Figure 2)
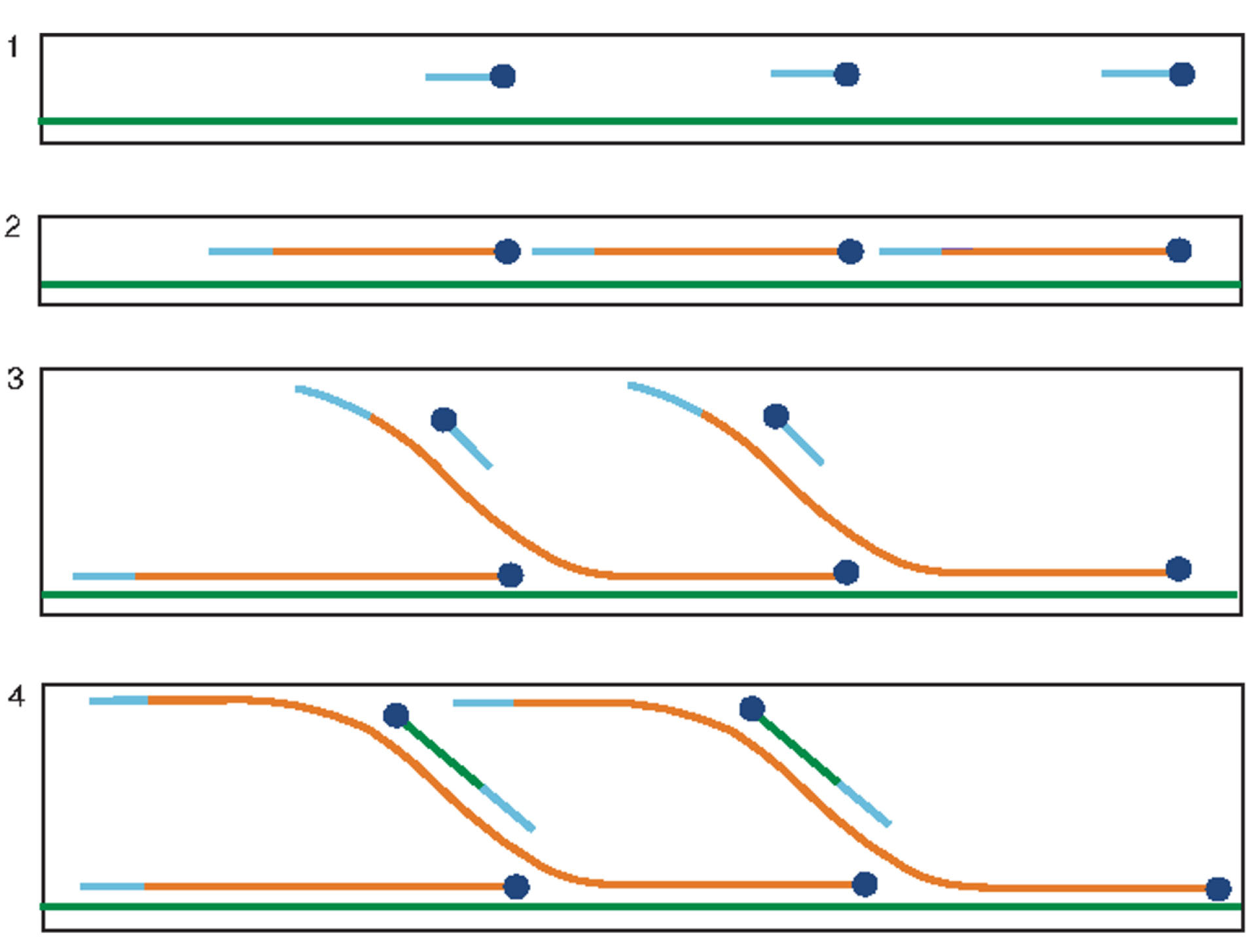
Figure 2.
Overview of the principle of MDA. 1) Random hexamers (blue lines) bind to ssDNA (green lines). 2) DNA synthesis is formed by Phi29 enzyme (blue circles) to create newly synthesized double-stranded DNA (dsDNA) (orange lines). 3) Primers bind to the newly synthesized DNA, and Phi29 displaces the strands and continues the polymerization. 4) Triggering polymerization on new strands leads to the formation of a hyperbranched structure. dNTP: deoxy nucleotide three phosphates. Adapted from Spits
71
with permission from Springer Nature, Copyright 2006.
.
Overview of the principle of MDA. 1) Random hexamers (blue lines) bind to ssDNA (green lines). 2) DNA synthesis is formed by Phi29 enzyme (blue circles) to create newly synthesized double-stranded DNA (dsDNA) (orange lines). 3) Primers bind to the newly synthesized DNA, and Phi29 displaces the strands and continues the polymerization. 4) Triggering polymerization on new strands leads to the formation of a hyperbranched structure. dNTP: deoxy nucleotide three phosphates. Adapted from Spits
71
with permission from Springer Nature, Copyright 2006.
MDA is an appropriate method for DNA amplification from small biological samples. This technique’s application is expanding due to particular advantages, including the capability to analyze defective DNA samples,
72
production of long DNA fragments for the analyzing molecular markers, lack of sensitivity to GC content, and self-limitation of MDA reaction.
72-74
The major application of this method relies on the diagnosis of TB. In this line, a study was performed to evaluate the new two-step PCR-MDA by combining the MDA method IS6110-specific PCR to detect TB infection. In this experiment, the MDA assay was used as an amplification step to increase the amount of DNA in clinical samples of saliva followed by IS6110 specific PCR. In this two-step assay, DNA polymerase, Phi29 was used to amplify the entire genome.
75
Helicase dependent amplification
HDA is an isothermal method to amplify DNA fragments at lower temperatures compared with PCR and uses the helicase enzyme to denature the DNA double helix (Figure 3).
76,77
HDA starts with the function of helicase enzyme separating DNA double helix and forming the ssDNA with DNA-binding proteins (ssBPs). Sequence-specific primers designed for the target DNA anneal to the template DNA’s borders during the third step. Then DNA polymerase extends the primers to generate new dsDNA. This reaction repeats for these new products.
78
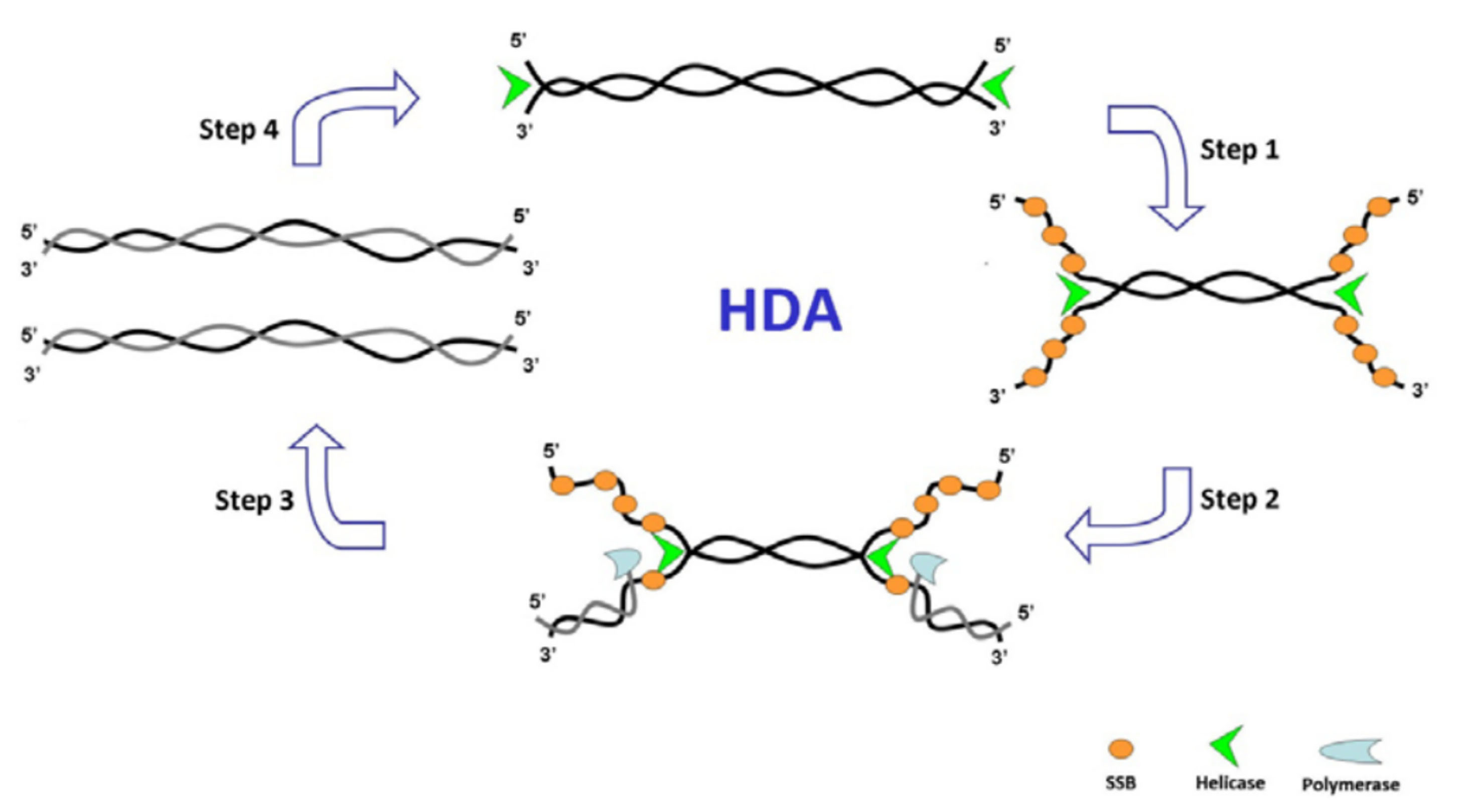
Figure 3.
Schematics of helicase dependent amplification. SSB: single-strand binding proteins. Adopted from Zanoliet al.
30
.
Schematics of helicase dependent amplification. SSB: single-strand binding proteins. Adopted from Zanoliet al.
30
The application of HDA is widespread for the detection of genetic polymorphisms to microbial tracking. In earlier attempts, HDA was used for the detection of Clostridium difficile due to its simplicity.
79
Later, it was used to rapidly detect S. aureus using a short DNA fragment specific to this microorganism.
80
Recently, Kolm et al
81
employed this method to detect the 16srRNA in Phylum bacteroides as water fecal pollution indicators. This marker is now known as “BacR” which was traditionally assessed by quantitative PCR (qPCR). The authors compared the newly designed HDA with the qPCR method and demonstrated that amplicon selection and primer design restricted the HDA performance. They also determined the limit of detection (LOD95%) for each technique and found that HDA performance was excellent compared to qPCR with 5.2-10.3 copies for HDA vs. 2.6-5.2 copies qPCR.
Loop-mediated isothermal amplification
Among different isothermal methods, LAMP ranks the first, as 60.7% of all the publications listed in Web of Science by 2019 are reported to use this method, followed by RCA (16.2%). LAMP is reputed for its robust and high analytical sensitivity and specificity to amplify target DNA. This is because this method uses up to six primers. However, this also increases the risk of forming primer dimers due to many long primers.
82
The isothermal and energy-efficient amplification requirements render LAMP as the first candidate for low-cost analysis and diagnostics at the point of need.
25
Another advantage of LAMP is its ability to be combined with the RT enzyme to use RNA as a template.
83
More importantly, LAMP assays are shown to excel RT-PCR performance with a ~100 times more sensitivity, capable of detecting viral RNA template down to 1–10 copies per reaction.
84
Mechanistically, LAMP employs the strand displacement principle and uses two pairs of specific primers to detect six different template DNA areas. It is described for the first time by Notomi et al.
85
In contrast to PCR reaction, DNA denaturation and primer annealing in LAMP occurs in a non-temperature-dependent manner, which relies on the kinetics of the reaction and the physical distance between the target sequence and primers in a loop.
86,87
The LAMP technique uses Bst DNA polymerase with the ability to mediate strand displacement reactions. The process is generally divided into two stages: the beginning of the structure production and the cycling reinforcement phase. In the first stage, all four primers are used, while in the second stage, only two internal primers are used. Single-stranded DNA is released by DNA synthesis.
26
The ssDNA released by an outer primer (F3) acts as a template for the next step of synthesis initiated by BIP and B3 and generates a stem-loop DNA structure.
88
Starting with a complimentary inner loop primer on the product will continue the cycling amplification process of each inner primer intermittently. This process is seen in a geometric aggregation of 109 copies of the target sequence in less than one hour.
89
The end products of stem-loop DNA can be distinguished by multiple repeats and cauliflower-like structures with multiple loops even using the real-time method and the endpoint method.
83
These steps are shown in detail in Figure 4. In this figure, (a) shows the primer design of the LAMP reaction. Six distinct regions are selected labeled F3, F2, F1, B1c, B2c, and B3 from the 5’end in which c represents complementary. The inner and outer primers used are FIP and BIP, and F3 and B3, respectively. (b) The process of synthesis starts from FIP. The F2 region binds to the F2c to initiate elongation. BIP similarly helps DNA amplification. The F3 binds to the F3c, and strand displacement DNA synthesis occurs. The elongated DNA from FIP is replaced and released. The resulted ss-DNA forms a loop in 5’ ends (structure 4).
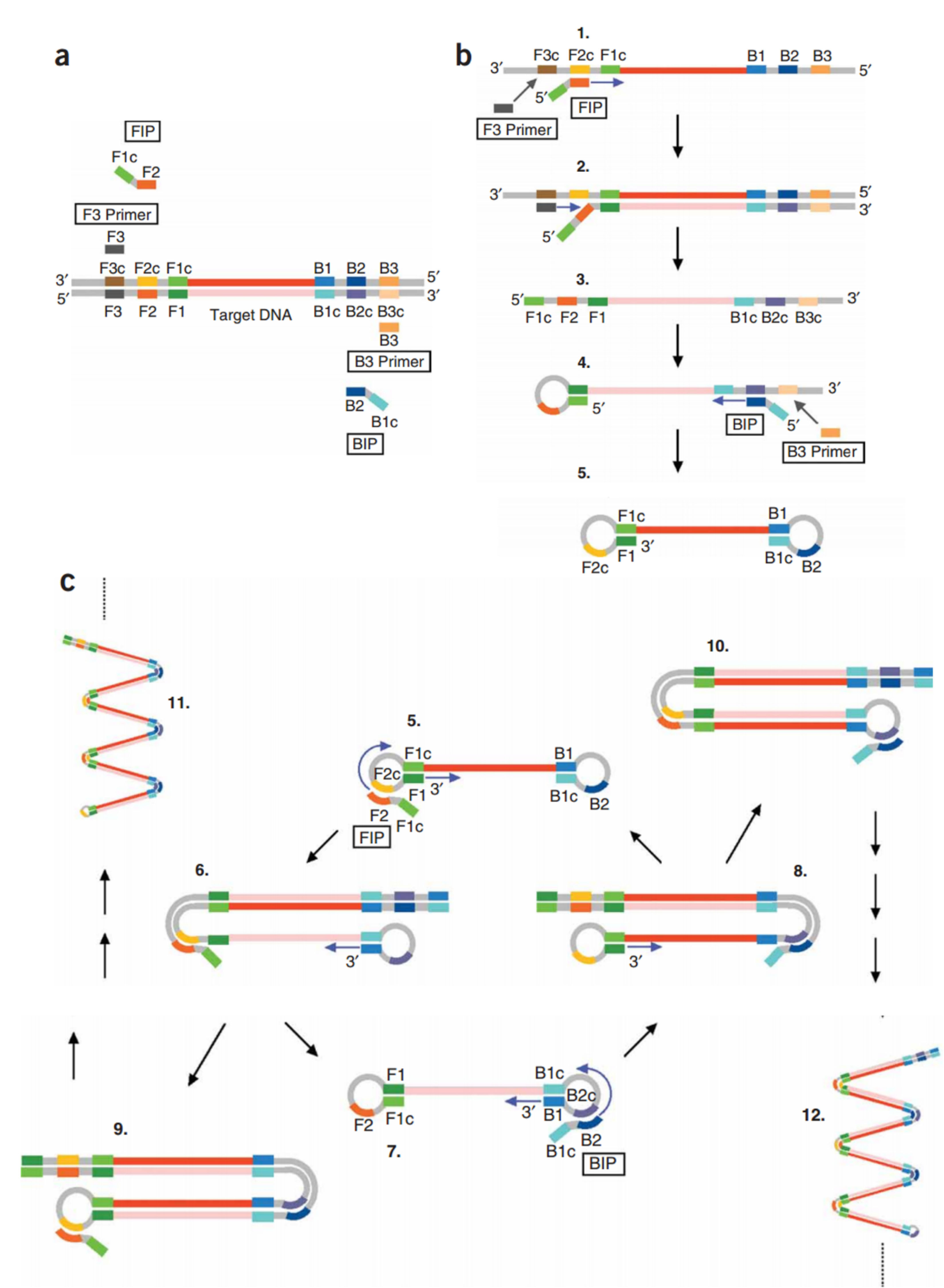
Figure 4.
Schematic representation of loop-mediated amplification. Adapted from Tomitaet al
90
with permission from Springer Nature Copyright 2008.
.
Schematic representation of loop-mediated amplification. Adapted from Tomitaet al
90
with permission from Springer Nature Copyright 2008.
DNA synthesis continues with the ss-DNA and BIP and B3 primers to generate structure 5 containing the loop structure at both ends (dumbbell-like structures). (c) Cycling amplification step. Structure 5 is used as a template, and self-primed DNA synthesis is started from the 3’ end of the F1 region and the annealing of FIP to single-stranded F2c region in the loop structure. After completion of the several steps, structure 7, complementary to structure 5, is generated. Structure 5 results from a similar reaction from structure eight, which originated from structures 5-7. Intermediate structures, including structures 7a and 9a, and structures 5a and 10a, are the products of structures 9 and 10, respectively. However, the displaced strands 7a and 5a are dumbbell-like structures 7 and 5, respectively. More elongated structures such as 11 and 12 are also generated.
90
As mentioned before, because of the large number of primers, the LAMP’s specificity and sensitivity are greatly increased. Therefore, it is considered a useful molecular method for detecting pathogens, such as MRSA in blood cultures. Equally, WHO published policy guidance for the use of LAMP for the diagnosis of pulmonary tuberculosis.
91
Because S. aureus is one of the most important pathogens that is causing deadly infections, its rapid and accurate diagnosis by the LAMP method has produced satisfying results.
92
A recent study by Foo et al on Entamoeba histolytica DNA samples reported better LOD results for LAMP and its rapidity in nucleic acid amplification compared with PCR, nested PCR, and real-time PCR. This report, along with previously mentioned studies, confirmed the LAMP technique’s advantages over the previous complex molecular methods in the rapid and sensitive detection of pathogenic microorganisms.
93
Nucleic acid sequence-based amplification
NASBA
94
that renowned as self-sustained sequence replication (3SR),
95
is known as a primer-dependent method that can utilize the continuous proliferation of nucleic acids in a single mixture at a constant temperature. This method is comparable to RT-PCR in that it is used to amplify single-stranded RNAs (ssRNAs). This method is based on the function of three enzymes: reverse transcription, RNase H, and DNA-dependent RNA polymerase. First, reverse transcriptase (RT) and RNase H transcripts complementary DNA (cDNA) from RNA. Then DNA-dependent RNA polymerase produces antisense RNAs from cDNA. In subsequent cycles, this antisense RNA and cDNA act as a template, and the cycle continue. Eventually, the accumulation of antisense RNA is created that is exactly like the original RNA. NASBA’s performance is far superior to that of RT-PCR, as NASBA can produce up to 109 times the RT-PCR of the product in 1.5 to 2 hours at 41°C.
96
Since the NASBA products are ssRNA; the denaturation step is omitted to detect hybridization, thus increasing the speed and ease of operations (Figure 5).
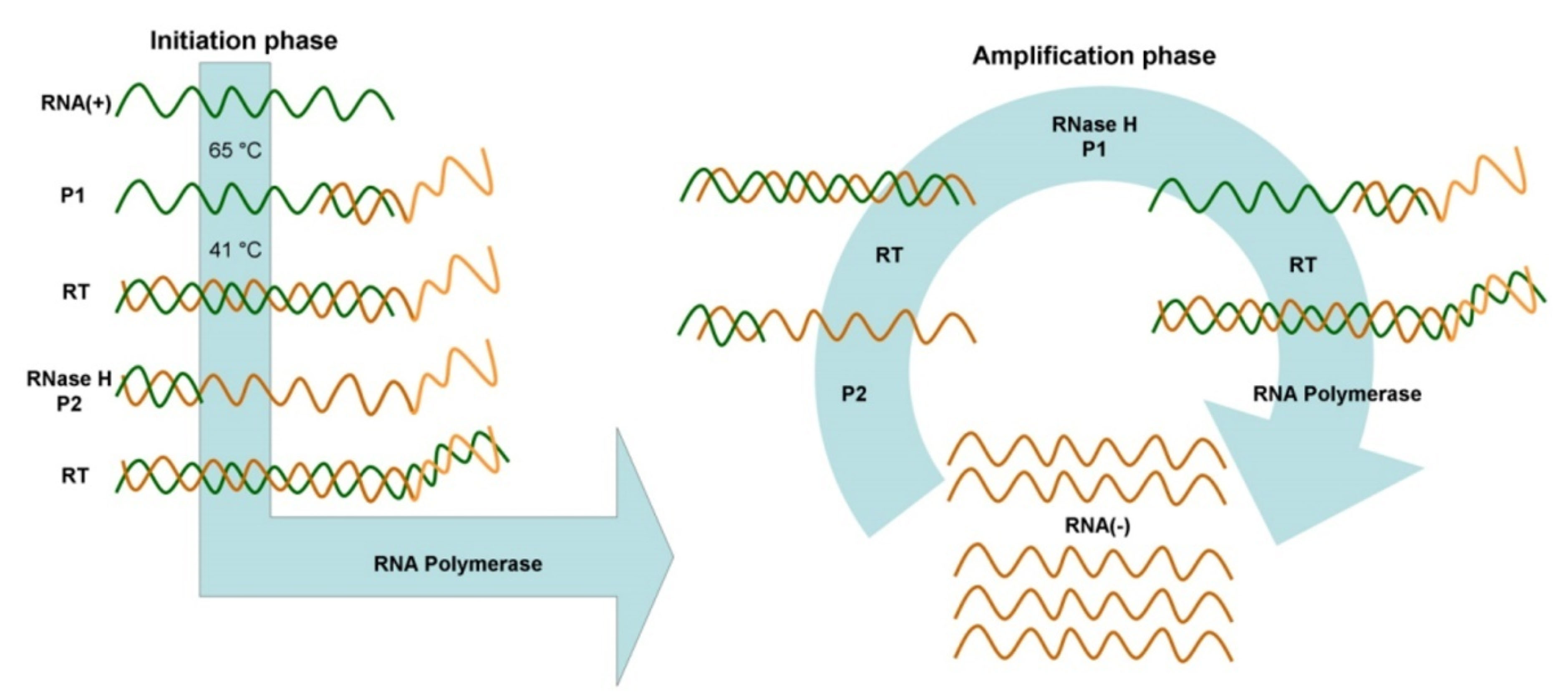
Figure 5.
Schematics of NASBA composed of two initiation and amplification cycles. Adopted from Zanoli et al.
30
.
Schematics of NASBA composed of two initiation and amplification cycles. Adopted from Zanoli et al.
30
The use of NASBA is extended to the rapid detection of bloodstream infections (BSI).
97
BSI is one of the leading causes of death in the US.
98,99
On the other hand, the inability of a rapid diagnosis fails to control the infection. Although various microorganisms can play a role in causing the disease, including Gram-positive bacteria and Gram-negative bacteria and various fungi, in most cases, BSI is associated with coagulase-negative Staphylococci (CoNS), S. aureus, Enterococcus spp., Candida species, and the Gram-negative bacilli including E. coli and Klebsiella spp.
100
The low sensitivity and time consuming of blood culture in BSI diagnosis have replaced the traditional culture-based techniques with newer methods such as NASBA. Also, interpreting the results of the identification of antibiotic-resistant bacteria in conventional methods is a time-consuming process.
101
Another method used for this purpose is real-time PCR, though the complexity and high cost limit its use. However, the accuracy of NASBA detection for blood pathogens and, at the same time, the need for less time to achieve optimal results have made NSABA a popular method. NASBA enabled identifying a wide range of microorganisms and is applied to detect specific genes on 16S rRNA bacteria and 28S rRNA fungi.
102,103
Transcription mediated amplification
As another isothermal amplification system, TMA also works without changing the temperature and requires both polymerase and reverse transcriptase enzymes to amplify nucleic acid. Rapid replication of the target RNA or DNA allows the simultaneous identification of different pathogenic organisms in a single tube (Figure 6). TMA technology enables clinical laboratories to perform a nucleic acid assay for blood screening with shorter process times and faster results.
104
This method is used in molecular biology and molecular medicine to identify and diagnose pathogenic organisms quickly. Contrary to similar techniques such as PCR and ligase chain reaction, this method for RNA transcription through RNA polymerase and DNA synthesis by reverse transcriptase can create a source RNA product or replication product from a target nucleic acid.
105
So, this method can be used to target both RNA and DNA. It has many advantages over other amplification methods: (i) it is isothermal; a heated block or a hot water bath is used instead of thermal cycler; (ii) TMA produces RNA replicons instead of DNA. Since RNA is more unstable in the laboratory environment than DNA, this advantage reduces the likelihood of excessive contamination, and (iii) TMA generates 100 to 1000 copies per cycle. This advantage results in an increased number of DNA/RNA copies by 10 billion times in about 15 to 30 minutes.
106
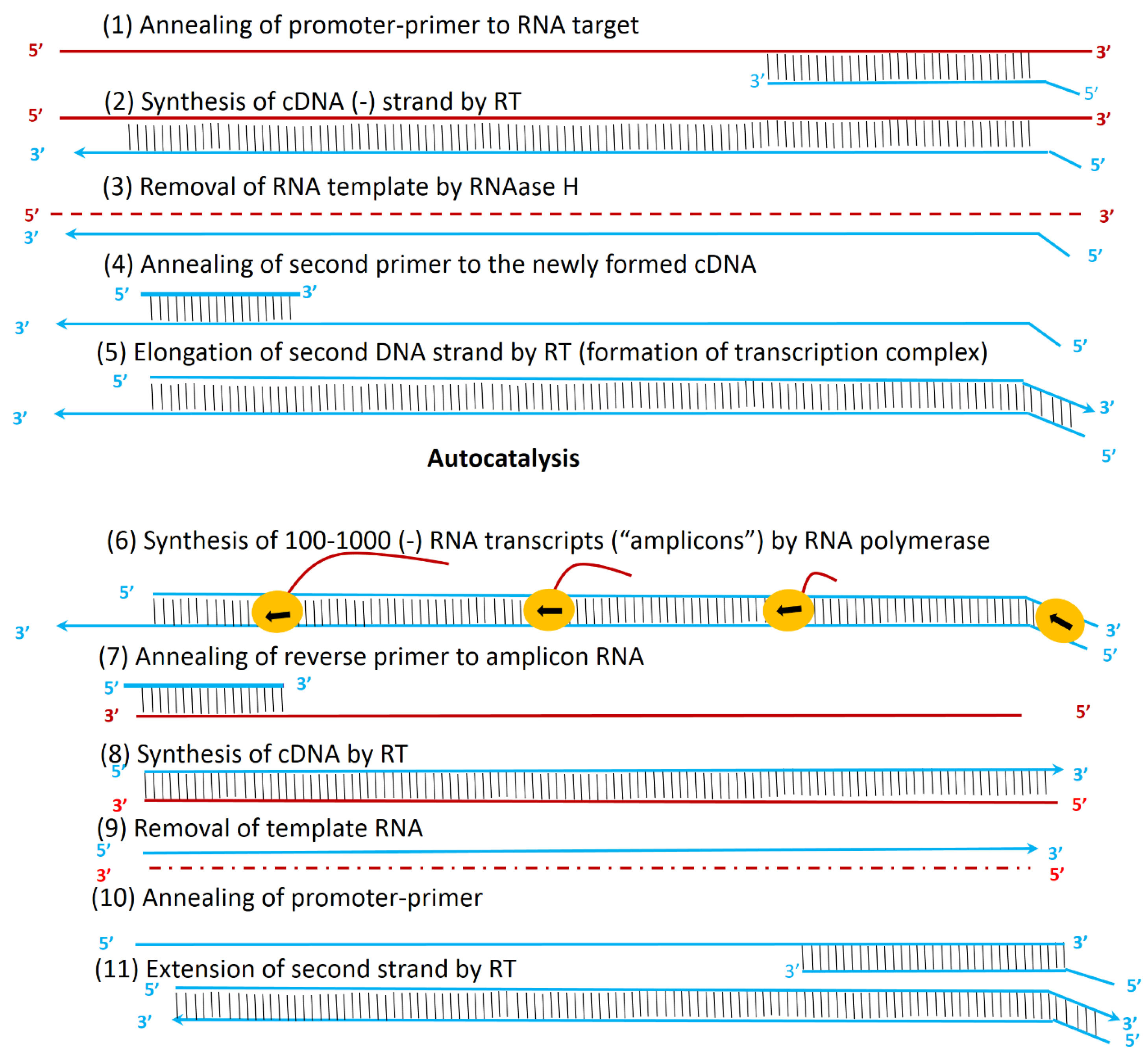
Figure 6.
Principle of TMA for the amplification of nucleic acids. Reprinted from Langabeer et al
106
with permission from Springer Nature Copyright 2002.
.
Principle of TMA for the amplification of nucleic acids. Reprinted from Langabeer et al
106
with permission from Springer Nature Copyright 2002.
Rolling circle amplification
RCA involves two types of hyperbranched rolling circle amplification (HRCA) and primer generation-rolling circle amplification (PG-RCA), which is collectively called exponential RCA.
107
RCA employs a padlock as a circular template synthesized from ssDNA and is responsible for producing a long and repetitive ssDNA.
108-110
Two types of enzymes are used in HRCA, including DNA polymerase for the extension of the sequence and DNA-ligase for the padlock probe’s circulation. The primers of this method are of two types; the first primer (P1) is complementary to the padlock probe while the second primer (P2) is paired with the ssDNA made from the padlock probe (Figure 7).
111
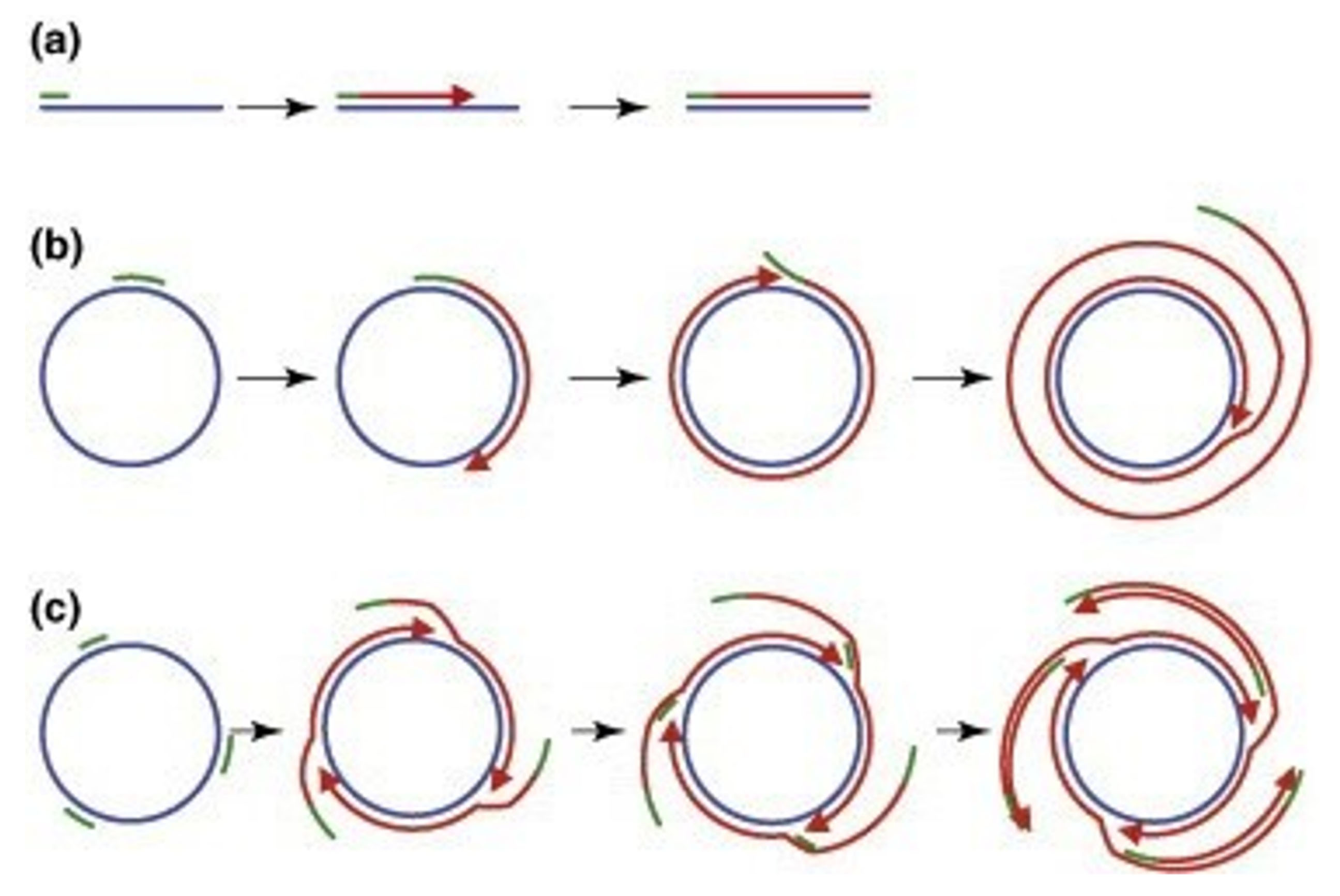
Figure 7.
Rolling-circle amplification of viral DNA genomes using Phi29 polymerase. Blue lines represent target DNA, green and red bars indicate oligonucleotide primers and newly synthesized DNA, respectively. Arrowheads demonstrate 3'ends of the growing DNA. (a) Primer binds to the template and a complementary strand is synthesized. (b) Single primer binds to the circular template and the new complementary strand is synthesized. After completion of one round, the synthesized strand and primer are displaced and a new round starts. The result is the production of several concatemeric ssDNA. (c) Multiple random primers bind to the circular template and DNA synthesis occurs in the same way as in b. The multiple products of this reaction are long concatemeric molecules of dsDNA. Reprinted from Johne et al
120
with permission from Elsevier Copyright 2009.
.
Rolling-circle amplification of viral DNA genomes using Phi29 polymerase. Blue lines represent target DNA, green and red bars indicate oligonucleotide primers and newly synthesized DNA, respectively. Arrowheads demonstrate 3'ends of the growing DNA. (a) Primer binds to the template and a complementary strand is synthesized. (b) Single primer binds to the circular template and the new complementary strand is synthesized. After completion of one round, the synthesized strand and primer are displaced and a new round starts. The result is the production of several concatemeric ssDNA. (c) Multiple random primers bind to the circular template and DNA synthesis occurs in the same way as in b. The multiple products of this reaction are long concatemeric molecules of dsDNA. Reprinted from Johne et al
120
with permission from Elsevier Copyright 2009.
This method’s basis is the strand replacement process leading to the continuous extension and eventually the creation of a segregated series of similar dsDNA fragments. Unlike HRCA, primer generation–rolling circle amplification (PG–RCA) employs nicking sites on the circular template instead of external primers. The nicking endonuclease (NEase) used in this reaction constantly generates these nicking sites to provide a 3́-OH end for DNA polymerase extension activity. The products of both HRCA and PG-RCA methods can be detected by gel electrophoresis and fluorescent dye-based assay.
111,112
In another study, RCA was used to identify the rpoB gene mutation in Mycobacterium tuberculosis.
113
The rpoB gene is responsible for encoding the β subunit of the bacterial RNA polymerase that contains the RIF resistance domain (RRDR).
114,115
Mutations in this gene cause multidrug-resistant strains.
116
Most RIF-resistant strains are also isoniazid-resistant bacteria. In this study, six lock-in probes were used to detect 12 common mutations in drug resistance in the RRDR rpoB gene from M. tuberculosis.
117
As another format of RCA, nick nuclease-mediated exponential RCA is applied for the fabrication of ultrasensitive biosensors. Taken as an example, in 2013, Du et al combined the RCA method with a terminal deoxynucleotidyl transferase (TdT) function. In this method, to improve the technique’s efficiency, the authors used DNase I to produce more -OH ends for the TdT function. During this method, RCA templates were prepared through the generation of dumbbell-like structures by the ligation of two hairpin DNA structures using a T4 ligase activity. The two loops of this dumbbell-like structure contained poly-T sequences acting as primer binding sites. Each loop contained a C-rich sequence to facilitate and improve the recognition of the RCA product. Consequently, the G-rich complementary sequences in RCA products can be folded to create a DNA secondary structure known as G-quadruplexes. Since the thioflavin T shows a precise response to G-quadruplexes, secondary structures play a key role in the detection step, and the RCA process can be tracked using a real-time PCR set.
118
Likewise, in a study by Jiang et al, authors developed nicking endonuclease-mediated exponential RCA for real-time monitoring of RCA. To this, they employed the specific fluorescence response of thioflavin T to G-quadruplex structures made by RCA products.
119
Strand displacement amplification
SDA technique introduced in 1992
121
is a DNA replication in which the transcription replication phase is omitted. As a rapid procedure, this isothermal method can produce about 107-fold amplification in 2 hours at 37°C.
122
SDA employs two types of enzymes: an endonuclease to create the specific 5’ and 3’ ends at the nicking site, and an exonuclease-deficient Klenow to extend the 3’ end. Following heating and denaturation by binding primers P1 and P2, two primer duplication patterns are created that give the 3’ end of the duplexes to expand polymerase and produce dsDNA containing nicking sites. These nicking sites are damaged by the endonuclease (NEase) tip. Following nicking, the newly generated 3’ ends at the nick sites initiate a new extension reaction with the displacement of downstream strand and then the generation of a new nicking site/reaction. The result of the nicking cycles and constant polymerization leads to the production of the target’s ss-compliment. The reaction employs the ss-complements from P1-T1 as the template for P2, while the ss-complements produced by P2-T2 act as the template for P1 (Figure 8).
122
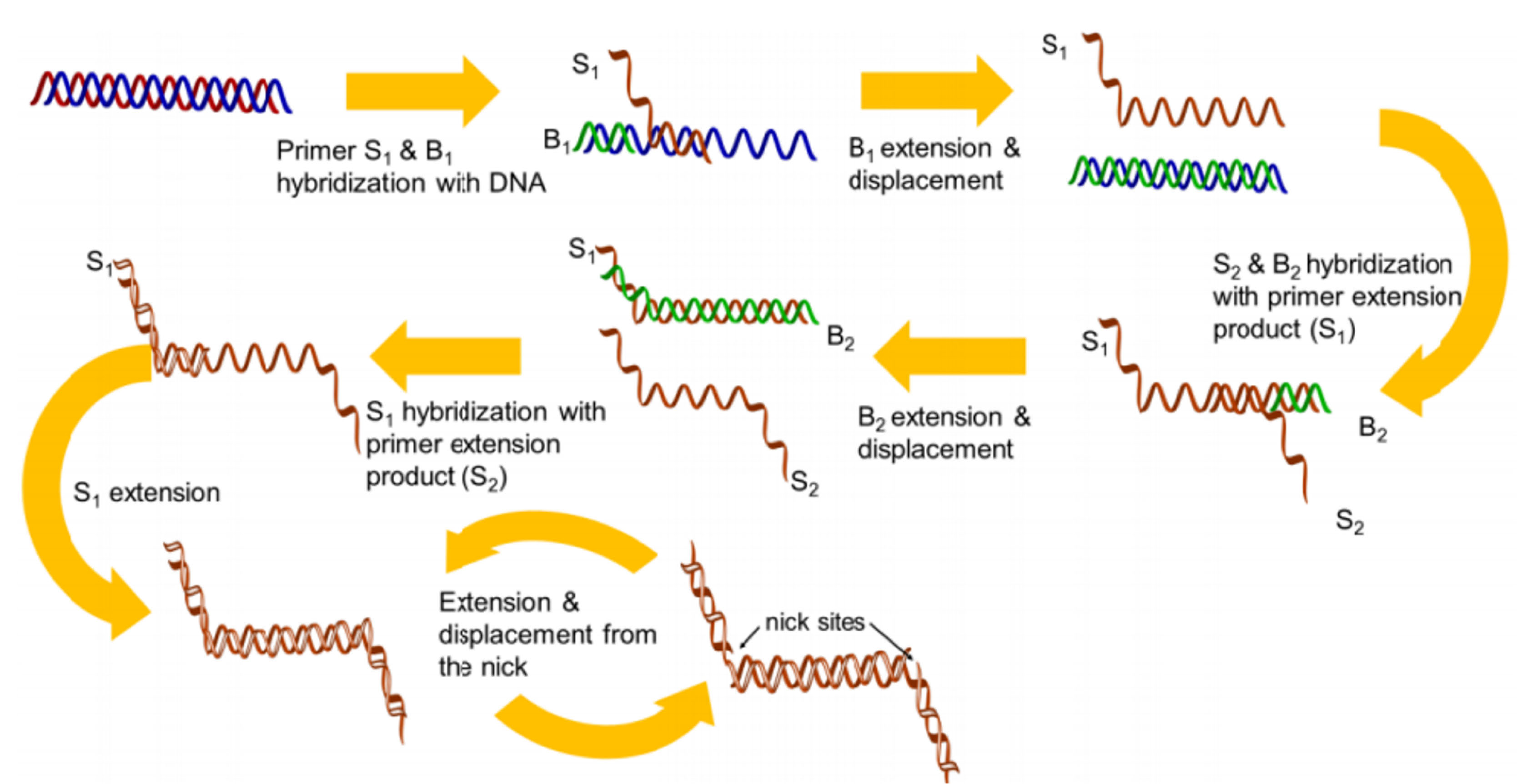
Figure 8.
Overview of SDA. Adopted from Chang et al.
123
.
Overview of SDA. Adopted from Chang et al.
123
The continuous repetition of these steps leads to the exponential accumulation of the target DNA. Today, S. aureus methicillin-resistant is a model for AMR determination using a novel gene insertion.
124
Regarding SDA’s specificity, an anchored system is reported for detecting methicillin resistance in S. aureus. In one study, the authors purified genomic DNA from cultured methicillin-resistant and sensitive S. aureus species and hybridized their DNA to the microelectronic chip array. Positive signals were only reported for MRSA isolates, where the human factor V gene was used as the control in the anchored SDA system. These results proved the specificity and accuracy of anchored SDA performance in methicillin-sensitive amplification reactions.
125,126
The advantages and disadvantages of above discussed methods are listed in Table 1.
Table 1.
Comparison of isothermal methods for amplification of nucleic acids
|
Isothermal method
|
Tm (◦C)
|
Time
|
Yield
|
Amplicon
|
Requirements
|
Primers
|
Template/ sensitivity
|
Ref.
|
| LAMP |
60 -65 |
15-60 min |
109-1010
|
Various amplicons in size (300 bp -10 kb) |
DNA Pol (Bst) |
3 pairs |
DNA/RNA |
82,89
|
| RPA |
37 -42 |
5-7min |
|
100-250 |
Recombinase (T4 UvsX), strand displacing DNA
Pol (POL I, Bsu)
|
1 pair |
DNA and RNA |
36-38,127
|
| RCA |
25–37 |
1h |
107-fold
|
Long ssDNA |
DNA Pol (Bst, ϕ29, vent (exo-), T7
RNA Pol)/ nicking endonuclease (NEase)
|
1 single primer and padlock probe |
circular ssDNA, RNA / microRNA |
111,127
|
| NASBA |
41 |
1.5h |
109-1012
|
RNA products |
DNA-dependent RNA pol (T7 RNA
Pol), RNase H, and (RT avian myeloblastosis virus (AMV) )
|
1 pair |
ssRNA, tmRNA, rRNA |
97,128
|
| HDA |
60-65 |
1-1.5h |
>106
|
<150bp |
thermostable helicase (Tte-UvrD)/
DNA Pol (Bst) or RT
|
1 pair |
DNA, rRNA |
77,81,129
|
| SDA |
37 |
2h |
107
|
- |
Exo- klenow, nicking endonucleases (HincII)
|
2 pair |
DNA |
122,130
|
| MDA |
30 |
2h |
- |
Producs larger than PCR ( > 70 kb) |
high processivity DNA Pol (ϕ29) |
Random hexamer |
dsDNA |
71,131
|
| TMA |
40-55 |
15-30 min |
1012
|
RNA products |
RNA polymerase (T7), RNase Hactivity of theRT
|
2 primers |
rRNA |
104,106
|
Abbreviations: polymerase (Pol); reverse transcriptase (RT); Endonuclease exonuclease-deficient klenow (exo-klenow).
Isothermal amplification methods integrated with nanotechnology and microfluidics
This section discusses the combination of isothermal amplification methods with two current advanced systems, namely nanoplatforms and lab-on-a-chip (LOC) microfluidic platforms.
Combination of isothermal methods with nanoplatforms (ultrasensitive biosensors
)
Nanoscale molecules with high potential to compute at a nano-sized level became much attractive to the researchers.
132
This is due to the special characteristic of DNA and RNA in recognition and hybridization to a complementary sequence, making them useful biomaterials to build the molecular logic gates.
133
DNA amplification methods, including PCR-based techniques and thermal procedures, are widely used in clinical applications, especially diagnostic approaches.
134
The specificity and sensitivity of these methods are significant. The exponential amplification of DNA/RNA as a core technology in modern clinical diagnostics is used in constructing DNA sensors, DNA nano-machines, and DNA sequencing.
107,135
With regards to the development of supersensitive POC devices, the unique spectral features of nanomaterials such as gold (Au NPs) nanoparticles, called surface plasmon resonance (SPR) can be exploited for signal amplification in the fabrication of optical biosensors in which the detection systems are based on recognition of specific DNA (RNA) target.
127
Further isothermal amplification of captured (target) nucleic acids and then signal amplification using inherent features of Au nanoparticles (SPR effect) as well as other (metallic and non-metallic (organic)) nanoparticles (metallic nanoparticles) realize a specific yet ultrasensitive DNA-based biosensing system for AMR detection.
136
Another merit is the regeneration of the sensor surface that permits repetitive uses (at least five cycles) with a shortened assay time.
137
In this line, LAMP combined with the SPR-DNA arrays is shown to increases the sensitivity and specificity of the method in the detection of MRSA. LAMP amplification was conducted from blood cocultures and sputum for 0.23 kbp and 0.18 kbp DNA fragments of mecA and femB genes, respectively. The detection limit of LAMP-SPR sensing was ten copies/µl with a good selectivity toward the MRSA.
137
Equally, rapid detection of the Panton-Valentine leukocidin (PVL) toxin of MRSA exploiting Au NPs in two hours with LOD of ~200 copies of genomic DNA is obtained.
138
Recently, LAMP combined with nanogold probe (AuNP) is reported to achieve detection sensitivity even below the food safety control (<100 CFU/g for S. aureus enterotoxin A gene (sea) detection). In this formulation, LAMP-AuNP could detect as low as 9.7 fg (3.2 sea copies), which were lower than PCR (97 fg or 32 sea copies). Using crude DNA lysates in food samples, LAMP-AuNP detected down to 10 CFU/g for milk and ground pork samples in 27 minutes achieving 97.5–100% accuracy, 97–100% specificity, and 100% sensitivity, superior to the culture method, and comparable to PCR but without a need for a thermal cycler. The culture method also detected 104 CFU/g and 10 CFU/mL for both samples, respectively in 5–7 days.
139
In one study, Li et al
140
designed a logic gate in combination with isothermal amplification of DNA based on the nicking endonuclease method. They improved a series of three-input logic gates. Unlike previously developed methods by Chen et al,
141
which was based on forming interaction, this method was combined with a cycle amplification. Therefore, it could amplify the sample’s low concentrations, leading to an improved detection limit to 100-folds. Another study performed by Xue et al
142
reported the detection of ultra-low concentrations of DNA targets. In this preparation, the authors developed a system by the combination of magnetic nanoparticles and exonuclease III (ExoIII)-induced cascade two-stage isothermal amplification. For the integration of target binding and signal transduction sequences, a capture hairpin probe was assembled on magnetic nanoparticles. After sensing the analyte nucleic acid, the hairpin probe could be opened and removed by the ExoIII performance, leading to the release of target DNA and the generation of free signal transduction sequence of capture hairpin probe for the continuation of the next circular reaction. The new DNA could be hybridized with a hairpin DNA probe with a partially caged G-quadruplex sequence inside and forms a duplex structure releasing an active G-quadruplex structure. Then the formed duplex domain is digested by the ExoIII, which leads to the recycling of the process and generation of numerous ZnPPIX/G-quadruplex supermolecular complexes. These products were detected by zinc (II)-protoporphyrin IX (ZnPPIX), which acted as the optical label for fluorescent detection. Accordingly, this platform provided a detection limit of 0.75 fM to distinguish mismatch DNA from perfectly matched target DNA, which offers great potential for the early detection and diagnosis of genetic disorders.
142
Later, Li et al
143
used a similar method to detect bis-phenol A (BPA) in drinking water and food packaging materials. They developed a label-free aptamer fluorescent assay based on RCA/ExoIII-combined cascade amplification program. BPA recognition and signal amplification required the duplex DNA probe design containing anti-BPA aptamer and the trigger sequence. Then, in BSA’s presence, the trigger probe can be released and initiates RCA reaction to start the primary amplification. The RCA products work as the initiator of the secondary amplification assisted by ExoIII accompanied by hairpin probes to generate a large amount of G-quadruplex in lantern-like structures. Lastly, the G-quadruplex lanterns were illuminated by ZNPPIX to detect fluorescent signals. This method’s most important advantages were excellent sensitivity with a detection limit of 5.4×10-17M and high specificity in detecting BPA. In a recent study, RCA was used to determine the concentration of S. aureus by colorimetric determination. In this method, linear padlock probes were used to track the target sequence so that these probes, after binding to DNA, were labeled with biotin and act as a primer for RCA. As a result, RCA products containing biotin labels were hybridized with digoxin-labeled signal probes, which were fixed on a plate. The change in color from colorless to blue due to the oxidation of tetramethylbenzidine by H2O2 made it possible to track the bacteria, resulting in 1.2 pM LOD for the target sequence of S. aureus (Figure 9).
112
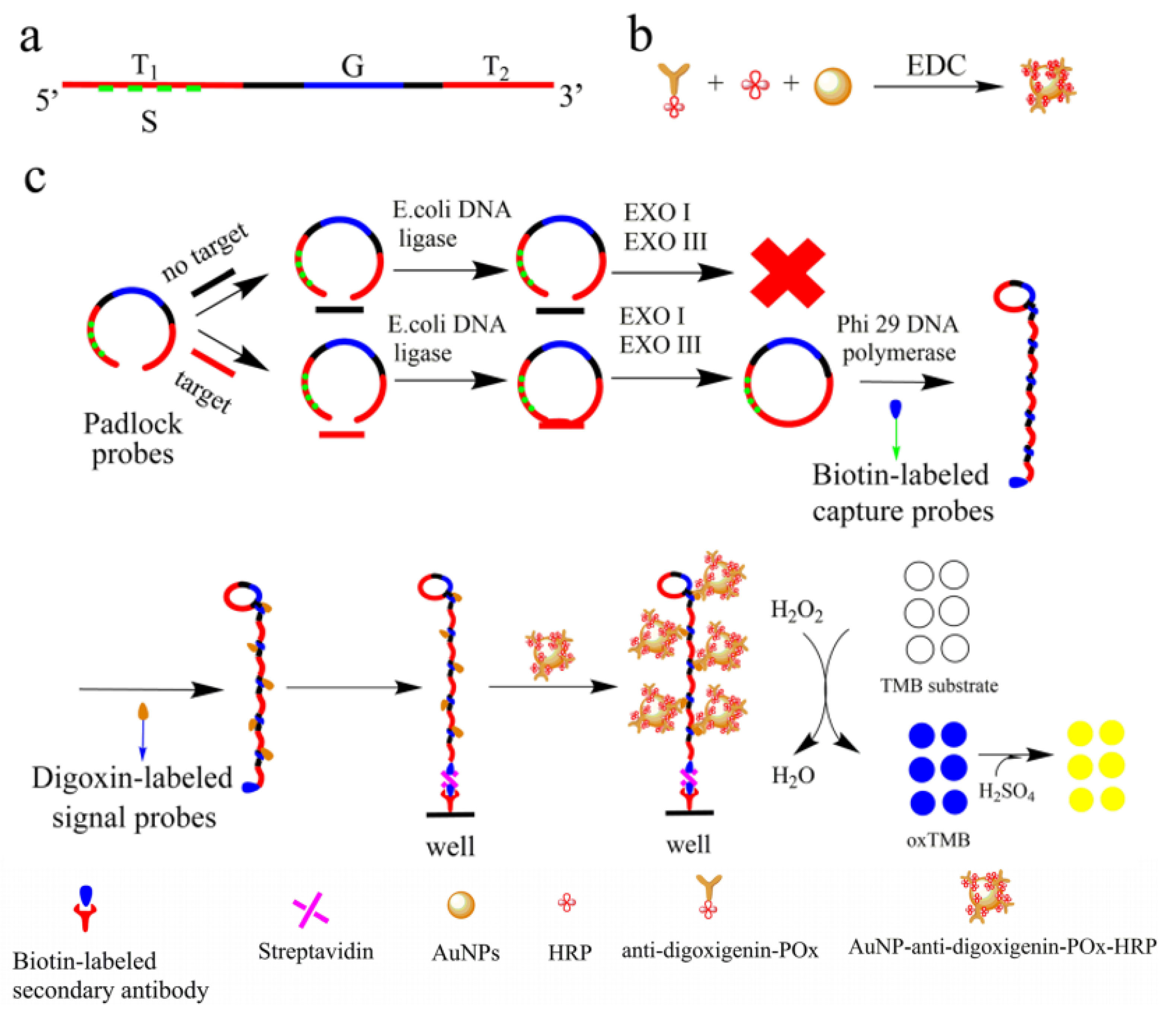
Figure 9.
Isothermal methods coupled with nanoplatforms. RCA amplification combined with gold NPs is used for signal amplification to detect AMR with ultra-sensitivity. Reprinted from Li et al.
112
with permission from Springer Nature Copyright 2020.
.
Isothermal methods coupled with nanoplatforms. RCA amplification combined with gold NPs is used for signal amplification to detect AMR with ultra-sensitivity. Reprinted from Li et al.
112
with permission from Springer Nature Copyright 2020.
In another study, Sedighi et al
144
integrated the HDA method with gold nano-particles to improve the helicase enzyme denaturation efficiency. This method’s advantages were improved denaturation, enhanced sensitivity, and specificity, and inhibited primer-dimer formation. Likewise, in an effort, Lin et al
145
conducted a digital LAMP or RT-LAMP on a commercially available membrane to eliminate the intricate chip fabrication need. The procedure involves the effective droplet formation on a commercial track-etched polycarbonate membrane, wherein each single DNA amplification reaction occurs in a pore which works as a nano-reactor. Authors were able to perform absolute quantification of bacterial genomic DNA with a dynamic range of 11-1.1×105 copies/µL. This method was also used for the quantification of MS2 bacteriophage in wastewater. The discrimination between positive and negative probes can be done easily with a 100 times difference in fluorescence intensities. This method can be applied as a low-cost, flexible, and simplified system for POC users or common laboratories.
In an additional setting, Khater et al
146
developed a label-free highly integrated with in situ RPA amplification and detection system needing room temperature to perform the reaction. This method uses the advantages of AuNPs-modified sensing substrates and electrochemical impedance spectroscopic detection. As the proof-of-concept, Citrus tristeza virus was detected by this system. The authors transferred the optimized RPA conditions to the AuNP-modified electrode surface previously modified with a thiolated forward primer. The Citrus tristeza virus target was amplified in situ and analyzed by EUS in a Fe (CN6)4-/Fe (CN6)3- red-ox system. This system is potentially able to detect 1000 fg/µL of nucleic acid. The lower cost, higher sensitivity, and portability made this system superior to the RPA in a homogeneous phase.
Notably, isothermal-based amplification methods combined with nanoplatforms are commercially available as test kits for the fast detection of pathogenic agents. In 2019, Thongphueak et al
147
developed a rapid test kit to identify Campylobacter spp.in meat products using LAMP combined with lateral flow dipstick (LFD) and AuNPs. To this, they designed LAMP primers using the conversed nucleic acid region of Campylobacter spp. The sensitivity of the LAMP-LFD and LAMP-AuNP assays were measured as 360 fg/µL. The specificity of the analysis was high due to the absence of any cross-reactions to L. monocytogenes, S. typhimurium, E. coli, B. cereus, P. aeruginosa, S. aureus, E. aerogenes, S. marcescens, V. parahaemolyticus, V. cholera, K. oxytoca,and C. diversus. This POC showed a sensitivity of 100%, specificity of 95%, and an accuracy of 96.67% for both LAMP-LFD and LAMP-AuNPs.
Combination of isothermal amplification methods with microfluidic devices (high-throughput screening arrays)
Medical diagnostics, environmental observations, and quality control of food products require nucleic acid replication techniques such as PCR. High-efficiency multiplexing techniques have been identified as advantageous in micro-formulation, especially in single-drop microfluidics. The fundamental principle of separating the analyzed sample into a number of identical micro volumes gives the possibility of individual manipulation.
148
Each analog droplet is isolated from the reaction chamber, which has all the reaction’s necessary components. Simultaneous reacting of thousands of drops results in the development of various new applications that were not identified using other techniques. That is, digital replication methods provide more sensitive and reliable measurements of nucleic acid values.
149
And are used for studying point mutations, copy number variations, and clonal replication in next-generation sequencing,
150
and antimicrobial susceptibility testing (AST).
151
As one, by incorporating an adaptable microfluidic design, a phenotypic AST system determines the of bacteria existence, classifies major classes of bacteria, detects polymicrobial samples, and identifies antimicrobial susceptibility directly from clinical samples (urine, blood cultures, and whole blood). It can also analyze polymicrobial samples at the single-cell level in 30 minutes. Through a pilot study of 25 clinical urine samples, this system demonstrated a sensitivity of 100% and specificity of 83.33% for pathogen classification and achieved 100% concordance for AST.
151
Furthermore, microfluidic devices provide the possibility of using a tiny system to meet the need for complicated and expensive thermal cycler devices to amplify nucleic acids.
152
Special PCR is one of these alternatives that involve microchannels contained different regions with fixed temperatures.
153,154
For the first time in 1998, Kopp et al
155
amplified a 176 bp fragment of the DNA gyrase gene of N. gonorrhoeae using this method. Further introduction of oscillating PCR simplified the digital microfluidics, as this method integrates the speed of the continuous flow PCR with the cycling flexibility of stationery chamber-based PCR.
156
The marriage of microfluidic devices with isothermal amplification methods to realize effective AMR biosensing platforms has been quite fruitful. With this in mind, and arrayed emulsion droplet microfluidic device has been used for digital LAMP analysis. In the sense of isothermal amplification, a microfluidic droplet array chip can be designed to execute the digital LAMP process by creating emulsion droplets, sorting them into a 30 × 8 droplet array, and executing LAMP across the 240 trapped and separated droplets (with a volume of 0.22 nL) after only 40 min of reaction at 56 °C. This design reported accurate quantification of nucleic acids across a dynamic range of 50 to 2.5 × 103 DNA copies per μL, with LOD down to a single DNA molecule.
157
For one, Tupik et al
158
designed the study based on microfluidic mixing with LAMP technique by soft lithography. Authors developed microfluidic devices for fluorescent detection of isothermal propagation of target DNA in a droplet-based microfluidic format as new digital amplification methods with a more reliable and sensitive measurement of nucleic acids. In another study, Lutz et al
159
developed an RPM microfluidic system to identify the mecA antibiotic resistance gene of S. aureus for the identification of less than a copy of this gene in <20 minutes.
Moreover, a combination of microfluidics with nucleic acid amplifications brings about an unprecedented opportunity to fabricate high throughput analytic systems saving both reagent, time, and above that can work for low numbers of RNA/DNA templates. Owing to high specificity and sensitivity, isothermal-based microfluidics systems are very beneficial for diagnostic applications at the time of bacterial or viral outbreaks. Such that, droplet-based microfluidics combined with LAMP is used as a sensitive biosensor for amplification of extracted RNA templates of the invA gene to detect S. typhimurium by simultaneously performing ∼106 LAMP-assisted amplification reactions in picoliter-sized droplets. Then, incubation of collected droplets in a thermocycler at 68 °C for 30 min and subsequent fluorescence emission for positive droplets were quantified. This system’s advantages were its simplicity in operation, high sensitivity (1 positive droplet per 250 CFU of S. typhimurium), specificity, rapidity, and above all, the high-throughput nature compared to well-established conventional methods. LOD for detecting pure S. typhimurium was 5000 CFU/mL in the sample or 25 RNA template/25 μL LAMP reaction cocktail compared to LOD of 5 × 105 CFU/mL of bacteria in the milk sample. This method showed remarkable higher specificity in distinguishing S. typhimorium inv(A) gene from two other prevalent bacterial contaminating milk, including S. flexneri and S. aureus (Figure 10).
160
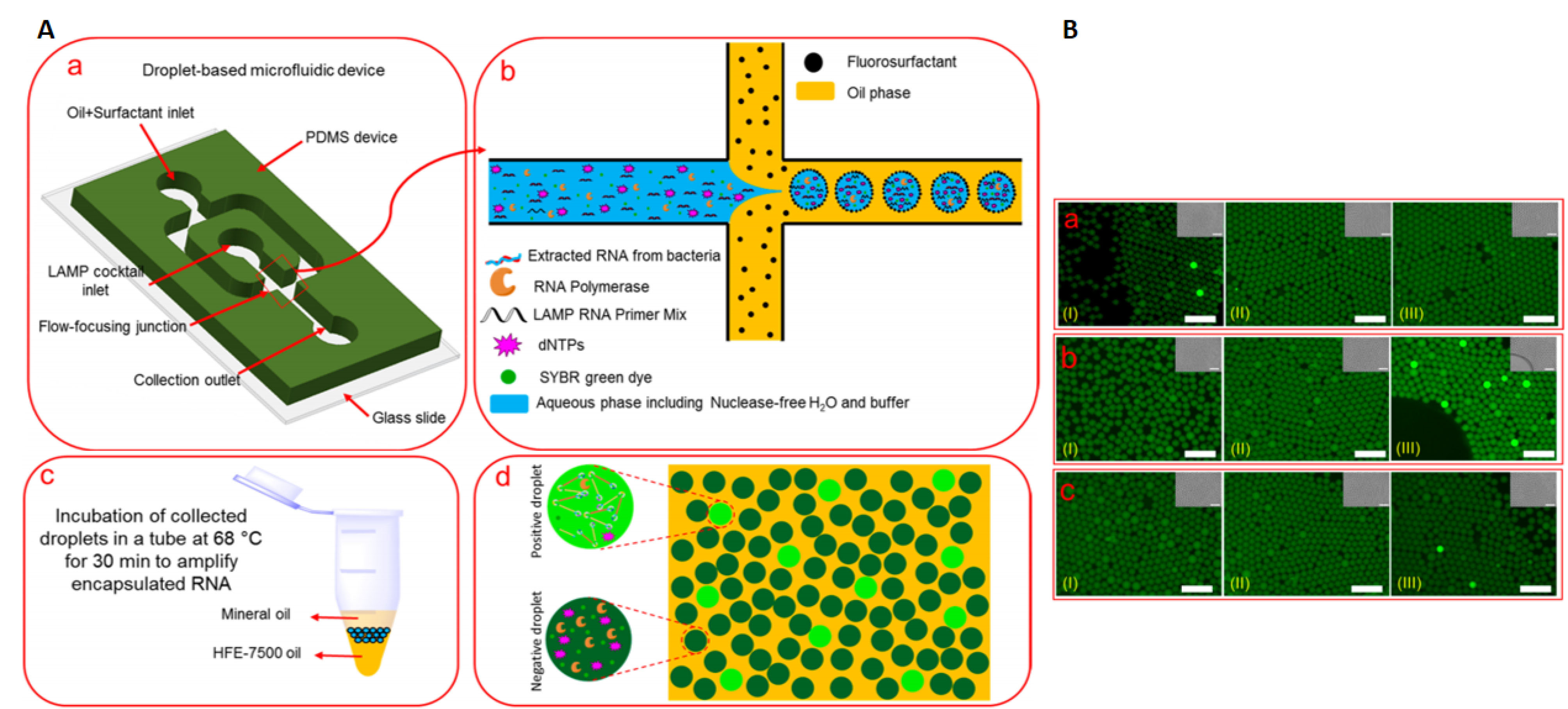
Figure 10.
LAMP-assisted droplet microfluidics. (A) Schematic representations of sample encapsulation, incubation, and screening. a, the LAMP reaction cocktail is encapsulated into water-in-oil droplets in the microfluidic flow-focusing device. b, Schematic cross-junction view of the flow-focusing, showing Collection of water-in-oil droplets into a PCR tube. c, Incubation of collected droplets (68°C for 30 min). d, Fluorescent imaging and "Yes/No" quantification of the incubated positive droplets. (B) Specificity of the LAMP-assisted droplet-based microfluidics sensor to detect the targeted bacteria. Reprinted from Azizi et al
160
with permission from American Chemical Society Copyright 2019.
.
LAMP-assisted droplet microfluidics. (A) Schematic representations of sample encapsulation, incubation, and screening. a, the LAMP reaction cocktail is encapsulated into water-in-oil droplets in the microfluidic flow-focusing device. b, Schematic cross-junction view of the flow-focusing, showing Collection of water-in-oil droplets into a PCR tube. c, Incubation of collected droplets (68°C for 30 min). d, Fluorescent imaging and "Yes/No" quantification of the incubated positive droplets. (B) Specificity of the LAMP-assisted droplet-based microfluidics sensor to detect the targeted bacteria. Reprinted from Azizi et al
160
with permission from American Chemical Society Copyright 2019.
Moreover, NASBA on a chip was demonstrated in 2004 by Gulliksen et al
161
by the real-time detection of a 1.0 mM human papillomavirus virus (HPV) in volumes as low as 10 nL in 41°C silicon-glass microchambers. These results were well compared to conventional NASBA with only a ratio of 1 to 2000 volumes of reaction and demonstrated the first records for the use of NASBA is a miniature identification system. Likewise, Ramalingam et al
162
designed a real-time HDA microfluidic device built on a glass-sealed PDMS that contained parallel 5 mL microchambers fed by microchannels from a single pipette loading port. Also, Burns et al
163
described a nanoliter silicon-glass apparatus to detect DNA using SDA and further electrophoresis at 17 minutes amplification time at 50 °C.
Surface-dependent ARCs were used by Sato et al
164
to develop a fully incorporated RCA microchip using hybrid padlock probes with 34 mm diameter beads as retainers of oligonucleotides and surfaces for RCA. A microchannel with a width of 210 mm and a depth of 40 mm, which holds a 0.5 ml of volume, was made on a glass with a cantilevered structure to hold the beads in place during the various stages of washing, injection of RCA solutions, and introduction of fluorescent probe for detection. The reaction temperature of 30°C and the performance of RCA microchips were demonstrated by Salmonella identification. In another attempt, Pardy et al
165
introduced a resistive heating alternative for disposable LoCNAAT devices. In this device, the author’s employed four-element samples with the different temperature dependence of resistivity profiles, which were by their favorable 60°C to 63°C temperature range. The temperature of sample BM117-83-B1 (one of the four used samples) reached the target temperature range for 5 minutes and was capable of holding this temperature for 25 minutes.
Moreover, a steady-state simulation analysis demonstrated that 85% of the reaction could keep the target temperature range. Eventually, the fabricated device was assessed using the LAMP assay to detect Chlamydia trachomatis template DNA. The same group
166
used a LoCNAAT method combined with LAMP to design a LOC instrument for isothermal amplification methods. The reaction was performed using a Bsm polymerase enzyme within the 20-25°C reaction temperature range. The device was set to sense C. trachomatis during an approximately 32-minute time. The advantages of this device were a smaller size, lower energy consumption, lower cost, and easily disposable.
Later, Pardy et al
167
provided a more completed device of the recent innovations, which was of low cost and wireless smart thermostat device for isothermal nucleic acid amplification as small as a 2 cm×3 cm LoC cartridge. It was equipped with Bluetooth connectivity, a 1200 mA battery, and a built-in OLED display and buttons for local status monitoring. The device provides 60ºC as the reaction temperature. When the LoC is filled with water, it takes 1.5 minutes for the heater and 6 minutes for the microreactor to reach this temperature (total 7 minutes). This device’s battery life was assessed and estimated a 3 hours, 15 minutes, and 55 seconds lifetime. The device supports isothermal nucleic acid amplification technology protocols with 35°C to 65°C range. The most important advantages of this prototype include full portability, the possibility of remote controlling and monitoring, and a fair price for good device performance.
Conclusion
Together, isothermal methods provide fast, available, portable, simple, and reliable tests for AMR detection and realize POC diagnostics to control infection, particularly in regions with limited sources/facilities. Furthermore improvements in isothermal amplification methods is achieved by their combination with other methods, such as nanotechnology, LOCs and microfluidics. This can pay the way for the development of rapid, available, ease of use miniature POC devices with high sensitivity, specificity, and multiplexing potential to detect simultaneous infections on a tiny paper strip/plastic chip. However, there are available commercial POCs, taken as an example glucose or blood pressure sensors, POCs for AMR detection are urgently needed and actively pursued.
Ethical Issues
Not applicable.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgments
This study was granted by the Tabriz University of Medical Sciences (TUMS).
References
-
National Institutes of Health (NIH). NIAID’s Antibacterial Resistance Program: Current Status and Future Directions. Washington, DC: NIH; 2014.
- Mendelson M, Matsoso MP. The World Health Organization Global Action Plan for antimicrobial resistance. S Afr Med J 2015; 105(5):325. doi: 10.7196/samj.9644 [Crossref] [ Google Scholar]
- Ji Y, Ma Z, Peppelenbosch MP, Pan Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob Health 2020; 8(4):e480. doi: 10.1016/s2214-109x(20)30068-1 [Crossref] [ Google Scholar]
- Kickbusch I, Leung G. Response to the emerging novel coronavirus outbreak. BMJ 2020; 368:m406. doi: 10.1136/bmj.m406 [Crossref] [ Google Scholar]
- Sun Q, Qiu H, Huang M, Yang Y. Lower mortality of COVID-19 by early recognition and intervention: experience from Jiangsu province. Ann Intensive Care 2020; 10(1):33. doi: 10.1186/s13613-020-00650-2 [Crossref] [ Google Scholar]
-
World Health Organization. High levels of antibiotic resistance found worlwide, new data shows. Availabe from: https://www.who.int/news/item/29-01-2018-high-levels-of-antibiotic-resistance-found-worldwide-new-data-shows. Accessed January 29, 2018.
- Law ILG, Loo JFC, Kwok HC, Yeung HY, Leung CCH, Hui M. Automated real-time detection of drug-resistant Mycobacterium tuberculosis on a lab-on-a-disc by recombinase polymerase amplification. Anal Biochem 2018; 544:98-107. doi: 10.1016/j.ab.2017.12.031 [Crossref] [ Google Scholar]
-
Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2019. CDC; 2019. 10.15620/cdc:82532
- Algammal AM, Hetta HF, Elkelish A, Alkhalifah DHH, Hozzein WN, Batiha GE. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist 2020; 13:3255-65. doi: 10.2147/idr.s272733 [Crossref] [ Google Scholar]
- Guo Y, Song G, Sun M, Wang J, Wang Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front Cell Infect Microbiol 2020; 10:107. doi: 10.3389/fcimb.2020.00107 [Crossref] [ Google Scholar]
- Khan MI, Xu S, Ali MM, Ali R, Kazmi A, Akhtar N. Assessment of multidrug resistance in bacterial isolates from urinary tract-infected patients. J Radiat Res Appl Sci 2020; 13(1):267-75. doi: 10.1080/16878507.2020.1730579 [Crossref] [ Google Scholar]
-
Antimicrobial resistance. WHO; 2020. Availabe from: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
- Strathdee SA, Davies SC, Marcelin JR. Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet 2020; 396(10257):1050-3. doi: 10.1016/s0140-6736(20)32063-8 [Crossref] [ Google Scholar]
- Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect 2020; 26(12):1622-9. doi: 10.1016/j.cmi.2020.07.016 [Crossref] [ Google Scholar]
- Jahrling PB. Microbe: are we ready for the next plague?. Emerg Infect Dis 2005; 11(11):1807. doi: 10.3201/eid1111.051084 [Crossref] [ Google Scholar]
- Walker GT, Quan J, Higgins SG, Toraskar N, Chang W, Saeed A. Predicting antibiotic resistance in gram-negative bacilli from resistance genes. Antimicrob Agents Chemother 2019; 63(4):e02462-18. doi: 10.1128/aac.02462-18 [Crossref] [ Google Scholar]
- Su M, Satola SW, Read TD. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 2019; 57(3):e01405-18. doi: 10.1128/JCM.01405-18 [Crossref] [ Google Scholar]
- Walker GT, Quan J, Higgins SG, Toraskar N, Chang W, Saeed A. Predicting antibiotic resistance in gram-negative bacilli from resistance genes. Antimicrob Agents Chemother 2019; 63(4):e02462-18. doi: 10.1128/AAC.02462-18 [Crossref] [ Google Scholar]
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020; 296(2):E32-E40. doi: 10.1148/radiol.2020200642 [Crossref] [ Google Scholar]
- Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 2020; 296(2):E115-E7. doi: 10.1148/radiol.2020200432 [Crossref] [ Google Scholar]
- Lan L, Xu D, Ye G, Xia C, Wang S, Li Y. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020; 323(15):1502-3. doi: 10.1001/jama.2020.2783 [Crossref] [ Google Scholar]
- Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology 2020; 296(2):E41-E5. doi: 10.1148/radiol.2020200343 [Crossref] [ Google Scholar]
- Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol 2013; 133(3):1-4. doi: 10.1038/jid.2013.1 [Crossref] [ Google Scholar]
- Yu M, He S, Wu D, Zhu H, Webster C. Examining the multi-scalar unevenness of high-quality healthcare resources distribution in China. Int J Environ Res Public Health 2019; 16(16):2813. doi: 10.3390/ijerph16162813 [Crossref] [ Google Scholar]
- Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, von Stetten F. Loop-mediated isothermal amplification (LAMP)–review and classification of methods for sequence-specific detection. Anal Methods 2020; 12(6):717-46. doi: 10.1039/c9ay02246e [Crossref] [ Google Scholar]
- Martzy R, Kolm C, Krska R, Mach RL, Farnleitner AH, Reischer GH. Challenges and perspectives in the application of isothermal DNA amplification methods for food and water analysis. Anal Bioanal Chem 2019; 411(9):1695-702. doi: 10.1007/s00216-018-1553-1 [Crossref] [ Google Scholar]
- Syal K, Mo M, Yu H, Iriya R, Jing W, Guodong S. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017; 7(7):1795-805. doi: 10.7150/thno.19217 [Crossref] [ Google Scholar]
- de Kraker ME, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050?. PLoS Med 2016; 13(11):e1002184. doi: 10.1371/journal.pmed.1002184 [Crossref] [ Google Scholar]
- García-Bernalt Diego J, Fernández-Soto P, Crego-Vicente B, Alonso-Castrillejo S, Febrer-Sendra B, Gómez-Sánchez A. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: towards a ready-to-use test. Sci Rep 2019; 9(1):14744. doi: 10.1038/s41598-019-51342-2 [Crossref] [ Google Scholar]
- Zanoli LM, Spoto G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors (Basel) 2013; 3(1):18-43. doi: 10.3390/bios3010018 [Crossref] [ Google Scholar]
- Duan R, Zuo X, Wang S, Quan X, Chen D, Chen Z. Lab in a tube: ultrasensitive detection of microRNAs at the single-cell level and in breast cancer patients using quadratic isothermal amplification. J Am Chem Soc 2013; 135(12):4604-7. doi: 10.1021/ja311313b [Crossref] [ Google Scholar]
- Lin Q, Ye X, Huang Z, Yang B, Fang X, Chen H. Graphene oxide-based suppression of nonspecificity in loop-mediated isothermal amplification enabling the sensitive detection of cyclooxygenase-2 mRNA in colorectal cancer. Anal Chem 2019; 91(24):15694-702. doi: 10.1021/acs.analchem.9b03861 [Crossref] [ Google Scholar]
- Treerattrakoon K, Jiemsakul T, Tansarawiput C, Pinpradup P, Iempridee T, Luksirikul P. Rolling circle amplification and graphene-based sensor-on-a-chip for sensitive detection of serum circulating miRNAs. Anal Biochem 2019; 577:89-97. doi: 10.1016/j.ab.2019.04.016 [Crossref] [ Google Scholar]
- Yuan K, Jiang Z, Jurado-Sánchez B, Escarpa A. Nano/micromotors for diagnosis and therapy of cancer and infectious diseases. Chemistry 2020; 26(11):2309-26. doi: 10.1002/chem.201903475 [Crossref] [ Google Scholar]
- Chen T, Ren L, Liu X, Zhou M, Li L, Xu J. DNA nanotechnology for cancer diagnosis and therapy. Int J Mol Sci 2018; 19(6). doi: 10.3390/ijms19061671 [Crossref]
- Lei R, Yan Z, Hu F, Zhu S, Xiong Y, Fan X. Rapid identification of quarantine invasive Solanum elaeagnifolium by real-time, isothermal recombinase polymerase amplification assay. RSC Adv 2017; 7(83):52573-80. doi: 10.1039/c7ra10781a [Crossref] [ Google Scholar]
- Patel P, Abd El Wahed A, Faye O, Prüger P, Kaiser M, Thaloengsok S. A field-deployable reverse transcription recombinase polymerase amplification assay for rapid detection of the chikungunya virus. PLoS Negl Trop Dis 2016; 10(9):e0004953. doi: 10.1371/journal.pntd.0004953 [Crossref] [ Google Scholar]
- Sun K, Xing W, Yu X, Fu W, Wang Y, Zou M. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of Schistosoma japonicum. Parasit Vectors 2016; 9(1):476. doi: 10.1186/s13071-016-1745-5 [Crossref] [ Google Scholar]
- Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol 2006; 4(7):e204. doi: 10.1371/journal.pbio.0040204 [Crossref] [ Google Scholar]
- Chao CC, Belinskaya T, Zhang Z, Ching WM. Development of recombinase polymerase amplification assays for detection of Orientia tsutsugamushi or Rickettsia typhi. PLoS Negl Trop Dis 2015; 9(7):e0003884. doi: 10.1371/journal.pntd.0003884 [Crossref] [ Google Scholar]
- Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Multiplexed recombinase polymerase amplification assay to detect intestinal protozoa. Anal Chem 2016; 88(3):1610-6. doi: 10.1021/acs.analchem.5b03267 [Crossref] [ Google Scholar]
- Boyle DS, McNerney R, Teng Low H, Leader BT, Pérez-Osorio AC, Meyer JC. Rapid detection of Mycobacterium tuberculosis by recombinase polymerase amplification. PLoS One 2014; 9(8):e103091. doi: 10.1371/journal.pone.0103091 [Crossref] [ Google Scholar]
- Crannell ZA, Castellanos-Gonzalez A, Irani A, Rohrman B, White AC, Richards-Kortum R. Nucleic acid test to diagnose cryptosporidiosis: lab assessment in animal and patient specimens. Anal Chem 2014; 86(5):2565-71. doi: 10.1021/ac403750z [Crossref] [ Google Scholar]
- Boyle DS, Lehman DA, Lillis L, Peterson D, Singhal M, Armes N. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio 2013; 4(2):e00135-13. doi: 10.1128/mBio.00135-13 [Crossref] [ Google Scholar]
- Natoli ME, Rohrman BA, De Santiago C, van Zyl GU, Richards-Kortum RR. Paper-based detection of HIV-1 drug resistance using isothermal amplification and an oligonucleotide ligation assay. Anal Biochem 2018; 544:64-71. doi: 10.1016/j.ab.2017.12.008 [Crossref] [ Google Scholar]
- Escadafal C, Paweska JT, Grobbelaar A, le Roux C, Bouloy M, Patel P. International external quality assessment of molecular detection of Rift Valley fever virus. PLoS Negl Trop Dis 2013; 7(5):e2244. doi: 10.1371/journal.pntd.0002244 [Crossref] [ Google Scholar]
- Euler M, Wang Y, Heidenreich D, Patel P, Strohmeier O, Hakenberg S. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents. J Clin Microbiol 2013; 51(4):1110-7. doi: 10.1128/jcm.02704-12 [Crossref] [ Google Scholar]
- Abd El Wahed A, El-Deeb A, El-Tholoth M, Abd El Kader H, Ahmed A, Hassan S. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PLoS One 2013; 8(8):e71642. doi: 10.1371/journal.pone.0071642 [Crossref] [ Google Scholar]
- Abd El Wahed A, Patel P, Heidenreich D, Hufert FT, Weidmann M. Reverse transcription recombinase polymerase amplification assay for the detection of Middle East respiratory syndrome coronavirus. PLoS Curr 2013; 5. doi: 10.1371/currents.outbreaks.62df1c7c75ffc96cd59034531e2e8364 [Crossref]
- Bonney LC, Watson RJ, Afrough B, Mullojonova M, Dzhuraeva V, Tishkova F. A recombinase polymerase amplification assay for rapid detection of Crimean-Congo haemorrhagic fever virus infection. PLoS Negl Trop Dis 2017; 11(10):e0006013. doi: 10.1371/journal.pntd.0006013 [Crossref] [ Google Scholar]
- Amer HM, Abd El Wahed A, Shalaby MA, Almajhdi FN, Hufert FT, Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J Virol Methods 2013; 193(2):337-40. doi: 10.1016/j.jviromet.2013.06.027 [Crossref] [ Google Scholar]
- Mo Y, Cui F, Li D, Dai Y, Li X, Zhang X. Establishment of a rapid and sensitive method based on recombinase polymerase amplification to detect mts90, a new molecular target of Mycobacterium tuberculosis. RSC Adv 2017; 7(79):49895-902. doi: 10.1039/c7ra09999a [Crossref] [ Google Scholar]
- Ng BYC, Wee EJH, Woods K, Anderson W, Antaw F, Tsang HZH. Isothermal point mutation detection: toward a first-pass screening strategy for multidrug-resistant tuberculosis. Anal Chem 2017; 89(17):9017-22. doi: 10.1021/acs.analchem.7b01685 [Crossref] [ Google Scholar]
- Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Mikrochim Acta 2014; 181(13-14):1715-23. doi: 10.1007/s00604-014-1198-5 [Crossref] [ Google Scholar]
- Toth IK, Bell KS, Holeva MC, Birch PR. Soft rot erwiniae: from genes to genomes. Mol Plant Pathol 2003; 4(1):17-30. doi: 10.1046/j.1364-3703.2003.00149.x [Crossref] [ Google Scholar]
- Riley MB, Williamson MR, Maloy O. Plant disease diagnosis. Plant Health Instr 2002. doi: 10.1094/phi-i-2002-1021-01 [Crossref]
- Ahmed FA, Larrea-Sarmiento A, Alvarez AM, Arif M. Genome-informed diagnostics for specific and rapid detection of Pectobacterium species using recombinase polymerase amplification coupled with a lateral flow device. Sci Rep 2018; 8(1):15972. doi: 10.1038/s41598-018-34275-0 [Crossref] [ Google Scholar]
- Dai T, Yang X, Hu T, Jiao B, Xu Y, Zheng X. Comparative evaluation of a novel recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay, LAMP, conventional PCR, and leaf-disc baiting methods for detection of Phytophthora sojae. Front Microbiol 2019; 10:1884. doi: 10.3389/fmicb.2019.01884 [Crossref] [ Google Scholar]
- Miao F, Zhang J, Li N, Chen T, Wang L, Zhang F. Rapid and sensitive recombinase polymerase amplification combined with lateral flow strip for detecting African swine fever virus. Front Microbiol 2019; 10:1004. doi: 10.3389/fmicb.2019.01004 [Crossref] [ Google Scholar]
- Zhao G, He H, Wang H. Use of a recombinase polymerase amplification commercial kit for rapid visual detection of Pasteurella multocida. BMC Vet Res 2019; 15(1):154. doi: 10.1186/s12917-019-1889-6 [Crossref] [ Google Scholar]
- Nelson MM, Waldron CL, Bracht JR. Rapid molecular detection of macrolide resistance. BMC Infect Dis 2019; 19(1):144. doi: 10.1186/s12879-019-3762-4 [Crossref] [ Google Scholar]
- Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother 1996; 40(8):1817-24. doi: 10.1128/aac.40.8.1817 [Crossref] [ Google Scholar]
- Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath AV. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol 1996; 22(5):867-79. doi: 10.1046/j.1365-2958.1996.01521.x [Crossref] [ Google Scholar]
- Banks DJ, Porcella SF, Barbian KD, Martin JM, Musser JM. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J Infect Dis 2003; 188(12):1898-908. doi: 10.1086/379897 [Crossref] [ Google Scholar]
- Giovanetti E, Brenciani A, Vecchi M, Manzin A, Varaldo PE. Prophage association of mef(A) elements encoding efflux-mediated erythromycin resistance in Streptococcus pyogenes. J Antimicrob Chemother 2005; 55(4):445-51. doi: 10.1093/jac/dki049 [Crossref] [ Google Scholar]
- Klaassen CH, Mouton JW. Molecular detection of the macrolide efflux gene: to discriminate or not to discriminate between mef(A) and mef(E). Antimicrob Agents Chemother 2005; 49(4):1271-8. doi: 10.1128/aac.49.4.1271-1278.2005 [Crossref] [ Google Scholar]
- Wang W, Ren Y, Lu Y, Xu Y, Crosby SD, Di Bisceglie AM. Template-dependent multiple displacement amplification for profiling human circulating RNA. Biotechniques 2017; 63(1):21-7. doi: 10.2144/000114566 [Crossref] [ Google Scholar]
- Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase Symmetrical mode of DNA replication. J Biol Chem 1989; 264(15):8935-40. [ Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A 2002; 99(8):5261-6. doi: 10.1073/pnas.082089499 [Crossref] [ Google Scholar]
- Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry 1988; 27(16):6008-13. doi: 10.1021/bi00416a027 [Crossref] [ Google Scholar]
- Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I. Whole-genome multiple displacement amplification from single cells. Nat Protoc 2006; 1(4):1965-70. doi: 10.1038/nprot.2006.326 [Crossref] [ Google Scholar]
- Panelli S, Damiani G, Espen L, Micheli G, Sgaramella V. Towards the analysis of the genomes of single cells: further characterisation of the multiple displacement amplification. Gene 2006; 372:1-7. doi: 10.1016/j.gene.2006.01.032 [Crossref] [ Google Scholar]
- Panelli S, Damiani G, Espen L, Sgaramella V. Ligation overcomes terminal underrepresentation in multiple displacement amplification of linear DNA. Biotechniques 2005; 39(2):174-80. doi: 10.2144/05392bm03 [Crossref] [ Google Scholar]
- Huang M, Yang F, Fu J, Xiao P, Tu J, Lu Z. Reaction parameter comparison and optimization of multiple displacement amplification. Anal Methods 2020; 12(1):46-53. doi: 10.1039/c9ay01922g [Crossref] [ Google Scholar]
- Wu N, Zhang Y, Fu J, Zhang R, Feng L, Hu Y. Performance assessment of a novel two-step multiple displacement amplification-PCR assay for detection of Mycobacterium tuberculosis complex in sputum specimens. J Clin Microbiol 2012; 50(4):1443-5. doi: 10.1128/jcm.05787-11 [Crossref] [ Google Scholar]
- Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep 2004; 5(8):795-800. doi: 10.1038/sj.embor.7400200 [Crossref] [ Google Scholar]
- Barreda-García S, Miranda-Castro R, de-Los-Santos-Álvarez N, Miranda-Ordieres AJ, Lobo-Castañón MJ. Helicase-dependent isothermal amplification: a novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal Bioanal Chem 2018; 410(3):679-93. doi: 10.1007/s00216-017-0620-3 [Crossref] [ Google Scholar]
-
Baker TA, Kornberg A. DNA Replication. 2nd ed. New York: WH Freeman and Company; 1992.
- Chow WH, McCloskey C, Tong Y, Hu L, You Q, Kelly CP. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J Mol Diagn 2008; 10(5):452-8. doi: 10.2353/jmoldx.2008.080008 [Crossref] [ Google Scholar]
- Frech GC, Munns D, Jenison RD, Hicke BJ. Direct detection of nasal Staphylococcus aureus carriage via helicase-dependent isothermal amplification and chip hybridization. BMC Res Notes 2012; 5:430. doi: 10.1186/1756-0500-5-430 [Crossref] [ Google Scholar]
- Kolm C, Martzy R, Führer M, Mach RL, Krska R, Baumgartner S. Detection of a microbial source tracking marker by isothermal helicase-dependent amplification and a nucleic acid lateral-flow strip test. Sci Rep 2019; 9(1):393. doi: 10.1038/s41598-018-36749-7 [Crossref] [ Google Scholar]
- Hardinge P, Murray JAH. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci Rep 2019; 9(1):7400. doi: 10.1038/s41598-019-43817-z [Crossref] [ Google Scholar]
- Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother 2009; 15(2):62-9. doi: 10.1007/s10156-009-0669-9 [Crossref] [ Google Scholar]
- Kashir J, Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med Hypotheses 2020; 141:109786. doi: 10.1016/j.mehy.2020.109786 [Crossref] [ Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000; 28(12):E63. doi: 10.1093/nar/28.12.e63 [Crossref] [ Google Scholar]
- Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev 2014; 27(4):783-822. doi: 10.1128/cmr.00003-14 [Crossref] [ Google Scholar]
- Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip 2012; 12(14):2469-86. doi: 10.1039/c2lc40100b [Crossref] [ Google Scholar]
- Seki M, Kilgore PE, Kim EJ, Ohnishi M, Hayakawa S, Kim DW. Loop-mediated isothermal amplification methods for diagnosis of bacterial meningitis. Front Pediatr 2018; 6:57. doi: 10.3389/fped.2018.00057 [Crossref] [ Google Scholar]
- Dolka B, Cisek AA, Szeleszczuk P. The application of the loop-mediated isothermal amplification (LAMP) method for diagnosing Enterococcus hirae-associated endocarditis outbreaks in chickens. BMC Microbiol 2019; 19(1):48. doi: 10.1186/s12866-019-1420-z [Crossref] [ Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 2008; 3(5):877-82. doi: 10.1038/nprot.2008.57 [Crossref] [ Google Scholar]
-
World Health Organization (WHO). The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. WHO; 2016.
- Misawa Y, Yoshida A, Saito R, Yoshida H, Okuzumi K, Ito N. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J Infect Chemother 2007; 13(3):134-40. doi: 10.1007/s10156-007-0508-9 [Crossref] [ Google Scholar]
- Foo PC, Nurul Najian AB, Muhamad NA, Ahamad M, Mohamed M, Yean Yean C. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: a comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol 2020; 20(1):34. doi: 10.1186/s12896-020-00629-8 [Crossref] [ Google Scholar]
- Compton J. Nucleic acid sequence-based amplification. Nature 1991; 350(6313):91-2. doi: 10.1038/350091a0 [Crossref] [ Google Scholar]
- Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD, Gingeras TR. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci U S A 1990; 87(5):1874-8. doi: 10.1073/pnas.87.5.1874 [Crossref] [ Google Scholar]
- Abd el-Galil KH, el-Sokkary MA, Kheira SM, Salazar AM, Yates MV, Chen W. Real-time nucleic acid sequence-based amplification assay for detection of hepatitis A virus. Appl Environ Microbiol 2005; 71(11):7113-6. doi: 10.1128/aem.71.11.7113-7116.2005 [Crossref] [ Google Scholar]
- Zhao Y, Park S, Kreiswirth BN, Ginocchio CC, Veyret R, Laayoun A. Rapid real-time nucleic acid sequence-based amplification-molecular beacon platform to detect fungal and bacterial bloodstream infections. J Clin Microbiol 2009; 47(7):2067-78. doi: 10.1128/jcm.02230-08 [Crossref] [ Google Scholar]
- Banerjee SN, Emori TG, Culver DH, Gaynes RP, Jarvis WR, Horan T. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989 National Nosocomial Infections Surveillance System. Am J Med 1991; 91(3b):86s-9s. doi: 10.1016/0002-9343(91)90349-3 [Crossref] [ Google Scholar]
- Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev 1993; 6(4):428-42. doi: 10.1128/cmr.6.4.428 [Crossref] [ Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004; 39(3):309-17. doi: 10.1086/421946 [Crossref] [ Google Scholar]
- Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 2004; 4(12):751-60. doi: 10.1016/s1473-3099(04)01205-8 [Crossref] [ Google Scholar]
- Simpkins SA, Chan AB, Hays J, Pöpping B, Cook N. An RNA transcription-based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett Appl Microbiol 2000; 30(1):75-9. doi: 10.1046/j.1472-765x.2000.00670.x [Crossref] [ Google Scholar]
- Widjojoatmodjo MN, Borst A, Schukkink RA, Box AT, Tacken NM, Van Gemen B. Nucleic acid sequence-based amplification (NASBA) detection of medically important Candida species. J Microbiol Methods 1999; 38(1-2):81-90. doi: 10.1016/s0167-7012(99)00079-2 [Crossref] [ Google Scholar]
-
Brentano ST, Mcdonough SH. Isothermal amplification of RNA by transcription-mediated amplification (TMA). In: Kessler C, ed. Nonradioactive Analysis of Biomolecules. Berlin, Heidelberg: Springer; 2000. p. 374-80. 10.1007/978-3-642-57206-7_31
- Wolk D, Mitchell S, Patel R. Principles of molecular microbiology testing methods. Infect Dis Clin North Am 2001; 15(4):1157-204. doi: 10.1016/s0891-5520(05)70190-2 [Crossref] [ Google Scholar]
- Langabeer SE, Gale RE, Harvey RC, Cook RW, Mackinnon S, Linch DC. Transcription-mediated amplification and hybridisation protection assay to determine BCR-ABL transcript levels in patients with chronic myeloid leukaemia. Leukemia 2002; 16(3):393-9. doi: 10.1038/sj.leu.2402392 [Crossref] [ Google Scholar]
- Seidi K, Neubauer HA, Moriggl R, Jahanban-Esfahlan R, Javaheri T. Tumor target amplification: implications for nano drug delivery systems. J Control Release 2018; 275:142-61. doi: 10.1016/j.jconrel.2018.02.020 [Crossref] [ Google Scholar]
- Daubendiek SL, Ryan K, Kool ET. Rolling-circle RNA synthesis: circular oligonucleotides as efficient substrates for T7 RNA polymerase. J Am Chem Soc 1995; 117(29):7818-9. doi: 10.1021/ja00134a032 [Crossref] [ Google Scholar]
- Fire A, Xu SQ. Rolling replication of short DNA circles. Proc Natl Acad Sci U S A 1995; 92(10):4641-5. doi: 10.1073/pnas.92.10.4641 [Crossref] [ Google Scholar]
- Liu D, Daubendiek SL, Zillman MA, Ryan K, Kool ET. Rolling circle DNA synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J Am Chem Soc 1996; 118(7):1587-94. doi: 10.1021/ja952786k [Crossref] [ Google Scholar]
- Murakami T, Sumaoka J, Komiyama M. Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res 2009; 37(3):e19. doi: 10.1093/nar/gkn1014 [Crossref] [ Google Scholar]
- Li Y, Wang J, Wang S, Wang J. Rolling circle amplification based colorimetric determination of Staphylococcus aureus. Mikrochim Acta 2020; 187(2):119. doi: 10.1007/s00604-019-4082-5 [Crossref] [ Google Scholar]
- Chen X, Wang B, Yang W, Kong F, Li C, Sun Z. Rolling circle amplification for direct detection of rpoB gene mutations in Mycobacterium tuberculosis isolates from clinical specimens. J Clin Microbiol 2014; 52(5):1540-8. doi: 10.1128/jcm.00065-14 [Crossref] [ Google Scholar]
- Mani C, Selvakumar N, Narayanan S, Narayanan PR. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J Clin Microbiol 2001; 39(8):2987-90. doi: 10.1128/jcm.39.8.2987-2990.2001 [Crossref] [ Google Scholar]
- Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 1998; 79(1):3-29. doi: 10.1054/tuld.1998.0002 [Crossref] [ Google Scholar]
- Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. J Clin Microbiol 2003; 41(5):2209-12. doi: 10.1128/jcm.41.5.2209-2212.2003 [Crossref] [ Google Scholar]
- Mitnick CD, Appleton SC, Shin SS. Epidemiology and treatment of multidrug resistant tuberculosis. Semin Respir Crit Care Med 2008; 29(5):499-524. doi: 10.1055/s-0028-1085702 [Crossref] [ Google Scholar]
- Du YC, Zhu YJ, Li XY, Kong DM. Amplified detection of genome-containing biological targets using terminal deoxynucleotidyl transferase-assisted rolling circle amplification. Chem Commun (Camb) 2018; 54(6):682-5. doi: 10.1039/c7cc09337c [Crossref] [ Google Scholar]
- Jiang HX, Liang ZZ, Ma YH, Kong DM, Hong ZY. G-quadruplex fluorescent probe-mediated real-time rolling circle amplification strategy for highly sensitive microRNA detection. Anal Chim Acta 2016; 943:114-22. doi: 10.1016/j.aca.2016.09.019 [Crossref] [ Google Scholar]
- Johne R, Müller H, Rector A, van Ranst M, Stevens H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol 2009; 17(5):205-11. doi: 10.1016/j.tim.2009.02.004 [Crossref] [ Google Scholar]
- Walker GT, Little MC, Nadeau JG, Shank DD. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc Natl Acad Sci U S A 1992; 89(1):392-6. doi: 10.1073/pnas.89.1.392 [Crossref] [ Google Scholar]
- Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res 1992; 20(7):1691-6. doi: 10.1093/nar/20.7.1691 [Crossref] [ Google Scholar]
- Chang CC, Chen CC, Wei SC, Lu HH, Liang YH, Lin CW. Diagnostic devices for isothermal nucleic acid amplification. Sensors (Basel) 2012; 12(6):8319-37. doi: 10.3390/s120608319 [Crossref] [ Google Scholar]
- Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 2006; 368(9538):874-85. doi: 10.1016/s0140-6736(06)68853-3 [Crossref] [ Google Scholar]
- Chang YC, Yang CY, Sun RL, Cheng YF, Kao WC, Yang PC. Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles. Sci Rep 2013; 3:1863. doi: 10.1038/srep01863 [Crossref] [ Google Scholar]
- Wang T, Zhang Z, Li Y, Xie G. Amplified electrochemical detection of mecA gene in methicillin-resistant Staphylococcus aureus based on target recycling amplification and isothermal strand-displacement polymerization reaction. Sens Actuators B Chem 2015; 221:148-54. doi: 10.1016/j.snb.2015.06.057 [Crossref] [ Google Scholar]
- Pumford EA, Lu J, Spaczai I, Prasetyo ME, Zheng EM, Zhang H. Developments in integrating nucleic acid isothermal amplification and detection systems for point-of-care diagnostics. Biosens Bioelectron 2020; 170:112674. doi: 10.1016/j.bios.2020.112674 [Crossref] [ Google Scholar]
- Ramezani R, Forouzandeh Moghadam M, Rasaee MJ. Development of sensitive and rapid RNA transcription-based isothermal amplification method for detection of Mycobacterium tuberculosis. Avicenna J Med Biotechnol 2019; 11(2):169-75. [ Google Scholar]
- Cao Y, Kim HJ, Li Y, Kong H, Lemieux B. Helicase-dependent amplification of nucleic acids. Curr Protoc Mol Biol 2013; 104:15.11.1-15. doi: 10.1002/0471142727.mb1511s104 [Crossref] [ Google Scholar]
- Zeng L, Mukama O, Lu X, Cao S, Lin D. Strand displacement amplification for multiplex detection of nucleic acids In: Singh A, Khan MW, eds Modulating Gene Expression-Abridging the RNAi and CRISPR-Cas9 Technologies. IntechOpen 2018. doi: 10.5772/intechopen.80687 [Crossref]
- Bleier S, Maier P, Allgayer H, Wenz F, Zeller WJ, Fruehauf S. Multiple displacement amplification enables large-scale clonal analysis following retroviral gene therapy. J Virol 2008; 82(5):2448-55. doi: 10.1128/jvi.00584-07 [Crossref] [ Google Scholar]
- Pischel U. Chemical approaches to molecular logic elements for addition and subtraction. Angew Chem Int Ed Engl 2007; 46(22):4026-40. doi: 10.1002/anie.200603990 [Crossref] [ Google Scholar]
- Frezza BM, Cockroft SL, Ghadiri MR. Modular multi-level circuits from immobilized DNA-based logic gates. J Am Chem Soc 2007; 129(48):14875-9. doi: 10.1021/ja0710149 [Crossref] [ Google Scholar]
- Jahanban-Esfahlan R, Seidi K, Jahanban-Esfahlan A, Jaymand M, Alizadeh E, Majdi H. Static DNA nanostructures for cancer theranostics: recent progress in design and applications. Nanotechnol Sci Appl 2019; 12:25-46. doi: 10.2147/nsa.s227193 [Crossref] [ Google Scholar]
- Jahanban-Esfahlan A, Seidi K, Jaymand M, Schmidt TL, Majdi H, Javaheri T. Dynamic DNA nanostructures in biomedicine: beauty, utility and limits. J Control Release 2019; 315:166-85. doi: 10.1016/j.jconrel.2019.10.003 [Crossref] [ Google Scholar]
- Nath P, Kabir A, Khoubafarin Doust S, Kreais ZJ, Ray A. Detection of bacterial and viral pathogens using photonic point-of-care devices. Diagnostics (Basel) 2020; 10(10):841. doi: 10.3390/diagnostics10100841 [Crossref] [ Google Scholar]
- Nawattanapaiboon K, Kiatpathomchai W, Santanirand P, Vongsakulyanon A, Amarit R, Somboonkaew A. SPR-DNA array for detection of methicillin-resistant Staphylococcus aureus (MRSA) in combination with loop-mediated isothermal amplification. Biosens Bioelectron 2015; 74:335-40. doi: 10.1016/j.bios.2015.06.038 [Crossref] [ Google Scholar]
- Yang AK, Lu H, Wu SY, Kwok HC, Ho HP, Yu S. Detection of Panton-Valentine leukocidin DNA from methicillin-resistant Staphylococcus aureus by resistive pulse sensing and loop-mediated isothermal amplification with gold nanoparticles. Anal Chim Acta 2013; 782:46-53. doi: 10.1016/j.aca.2013.04.004 [Crossref] [ Google Scholar]
- Srimongkol G, Ditmangklo B, Choopara I, Thaniyavarn J, Dean D, Kokpol S. Rapid colorimetric loop-mediated isothermal amplification for hypersensitive point-of-care Staphylococcus aureus enterotoxin A gene detection in milk and pork products. Sci Rep 2020; 10(1):7768. doi: 10.1038/s41598-020-64710-0 [Crossref] [ Google Scholar]
- Li X, Ding T, Sun L, Mao C. Ultrasensitive DNA detection by cycle isothermal amplification based on nicking endonuclease and its application to logic gates. Biosens Bioelectron 2011; 30(1):241-8. doi: 10.1016/j.bios.2011.09.019 [Crossref] [ Google Scholar]
- Chen X, Wang Y, Liu Q, Zhang Z, Fan C, He L. Construction of molecular logic gates with a DNA-cleaving deoxyribozyme. Angew Chem Int Ed Engl 2006; 45(11):1759-62. doi: 10.1002/anie.200502511 [Crossref] [ Google Scholar]
- Xue Q, Lv Y, Zhang Y, Xu S, Li R, Yue Q. Ultrasensitive fluorescence detection of nucleic acids using exonuclease III-induced cascade two-stage isothermal amplification-mediated zinc (II)-protoporphyrin IX/G-quadruplex supramolecular fluorescent nanotags. Biosens Bioelectron 2014; 61:351-6. doi: 10.1016/j.bios.2014.05.047 [Crossref] [ Google Scholar]
- Li X, Song J, Xue QW, You FH, Lu X, Kong YC. A label-free and sensitive fluorescent qualitative assay for bisphenol A based on rolling circle amplification/exonuclease III-combined cascade amplification. Nanomaterials (Basel) 2016; 6(10):190. doi: 10.3390/nano6100190 [Crossref] [ Google Scholar]
- Sedighi A, Oberc C, Whitehall V, Li PCH. NanoHDA: a nanoparticle-assisted isothermal amplification technique for genotyping assays. Nano Res 2017; 10(1):12-21. doi: 10.1007/s12274-016-1262-z [Crossref] [ Google Scholar]
- Lin X, Huang X, Urmann K, Xie X, Hoffmann MR. Digital loop-mediated isothermal amplification on a commercial membrane. ACS Sens 2019; 4(1):242-9. doi: 10.1021/acssensors.8b01419 [Crossref] [ Google Scholar]
- Khater M, de la Escosura-Muñiz A, Altet L, Merkoçi A. In situ plant virus nucleic acid isothermal amplification detection on gold nanoparticle-modified electrodes. Anal Chem 2019; 91(7):4790-6. doi: 10.1021/acs.analchem.9b00340 [Crossref] [ Google Scholar]
- Thongphueak D, Chansiri K, Sriyapai T, Areekit S, Santiwatanakul S, Wangroongsarb P. Development of the rapid test kit for the identification of Campylobacter spp Based on loop-mediated isothermal amplification (LAMP) in combination with a lateral flow dipstick (LFD) and gold nano-DNA probe (AuNPs). Sci Technol Asia 2019; 24(1):63-71. doi: 10.14456/scitechasia.2019.7 [Crossref] [ Google Scholar]
- Ayoubi-Joshaghani MH, Dianat-Moghadam H, Seidi K, Jahanban-Esfahalan A, Zare P, Jahanban-Esfahlan R. Cell-free protein synthesis: the transition from batch reactions to minimal cells and microfluidic devices. Biotechnol Bioeng 2020; 117(4):1204-29. doi: 10.1002/bit.27248 [Crossref] [ Google Scholar]
- Kiss MM, Ortoleva-Donnelly L, Beer NR, Warner J, Bailey CG, Colston BW. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal Chem 2008; 80(23):8975-81. doi: 10.1021/ac801276c [Crossref] [ Google Scholar]
- Abate AR, Hung T, Sperling RA, Mary P, Rotem A, Agresti JJ. DNA sequence analysis with droplet-based microfluidics. Lab Chip 2013; 13(24):4864-9. doi: 10.1039/c3lc50905b [Crossref] [ Google Scholar]
- Li H, Torab P, Mach KE, Surrette C, England MR, Craft DW. Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing. Proc Natl Acad Sci U S A 2019; 116(21):10270-9. doi: 10.1073/pnas.1819569116 [Crossref] [ Google Scholar]
- Gorgannezhad L, Stratton H, Nguyen NT. Microfluidic-based nucleic acid amplification systems in microbiology. Micromachines (Basel) 2019; 10(6):408. doi: 10.3390/mi10060408 [Crossref] [ Google Scholar]
- Ahrberg CD, Manz A, Chung BG. Polymerase chain reaction in microfluidic devices. Lab Chip 2016; 16(20):3866-84. doi: 10.1039/c6lc00984k [Crossref] [ Google Scholar]
- Polini A, Mele E, Sciancalepore AG, Girardo S, Biasco A, Camposeo A. Reduction of water evaporation in polymerase chain reaction microfluidic devices based on oscillating-flow. Biomicrofluidics 2010; 4(3):036502. doi: 10.1063/1.3481776 [Crossref] [ Google Scholar]
- Kopp MU, Mello AJ, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science 1998; 280(5366):1046-8. doi: 10.1126/science.280.5366.1046 [Crossref] [ Google Scholar]
- Zhang C, Wang H, Xing D. Multichannel oscillatory-flow multiplex PCR microfluidics for high-throughput and fast detection of foodborne bacterial pathogens. Biomed Microdevices 2011; 13(5):885-97. doi: 10.1007/s10544-011-9558-y [Crossref] [ Google Scholar]
- Ma YD, Luo K, Chang WH, Lee GB. A microfluidic chip capable of generating and trapping emulsion droplets for digital loop-mediated isothermal amplification analysis. Lab Chip 2018; 18(2):296-303. doi: 10.1039/c7lc01004d [Crossref] [ Google Scholar]
- Tupik A, Posmitnaya Y, Rudnitskaya G, Varlamov D, Evstrapov A. Microfluidic devices for loop mediated isothermal amplification. J Phys Conf Ser 2018; 1135(1):012110. doi: 10.1088/1742-6596/1135/1/012110 [Crossref] [ Google Scholar]
- Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Müller C. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA). Lab Chip 2010; 10(7):887-93. doi: 10.1039/b921140c [Crossref] [ Google Scholar]
- Azizi M, Zaferani M, Cheong SH, Abbaspourrad A. Pathogenic bacteria detection using RNA-based loop-mediated isothermal-amplification-assisted nucleic acid amplification via droplet microfluidics. ACS Sens 2019; 4(4):841-8. doi: 10.1021/acssensors.8b01206 [Crossref] [ Google Scholar]
- Gulliksen A, Solli L, Karlsen F, Rogne H, Hovig E, Nordstrøm T, Sirevåg R. Real-time nucleic acid sequence-based amplification in nanoliter volumes. Anal Chem 2004; 76(1):9-14. doi: 10.1021/ac034779h [Crossref] [ Google Scholar]
- Ramalingam N, San TC, Kai TJ, Mak MYM, Gong HQ. Microfluidic devices harboring unsealed reactors for real-time isothermal helicase-dependent amplification. Microfluid Nanofluidics 2009; 7(3):325. doi: 10.1007/s10404-008-0378-1 [Crossref] [ Google Scholar]
- Burns MA, Johnson BN, Brahmasandra SN, Handique K, Webster JR, Krishnan M. An integrated nanoliter DNA analysis device. Science 1998; 282(5388):484-7. doi: 10.1126/science.282.5388.484 [Crossref] [ Google Scholar]
- Sato K, Tachihara A, Renberg B, Mawatari K, Sato K, Tanaka Y. Microbead-based rolling circle amplification in a microchip for sensitive DNA detection. Lab Chip 2010; 10(10):1262-6. doi: 10.1039/b927460j [Crossref] [ Google Scholar]
- Pardy T, Tulp I, Kremer C, Rang T, Stewart R. Integrated self-regulating resistive heating for isothermal nucleic acid amplification tests (NAAT) in lab-on-a-chip (LoC) devices. PLoS One 2017; 12(12):e0189968. doi: 10.1371/journal.pone.0189968 [Crossref] [ Google Scholar]
- Pardy T, Rang T, Tulp I. Thermal analysis of a disposable, instrument-free DNA amplification lab-on-a-chip platform. Sensors (Basel) 2018; 18(6):1812. doi: 10.3390/s18061812 [Crossref] [ Google Scholar]
- Pardy T, Sink H, Koel A, Rang T. Development of a low-cost, wireless smart thermostat for isothermal DNA amplification in lab-on-a-chip devices. Micromachines (Basel) 2019; 10(7):437. doi: 10.3390/mi10070437 [Crossref] [ Google Scholar]