Advanced pharmaceutical bulletin. 15(2):453-466.
doi: 10.34172/apb.025.43819
Original Article
Silencing Calumenin Expression via Artificial MicroRNA, a Potential Breakthrough for Inhibiting Proliferation, Halting Migration, and Triggering Apoptosis in Breast Cancer Cells
Zahra Amiri Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, 1 
Fatemeh Bahrami Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, 1
Babak Jahangiri Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing, 1 
Arash Javeri Conceptualization, Methodology, 2
Frouzandeh Mahjoubi Conceptualization, Methodology, 3
Nahid Nafissi Resources, 4
Mohammad Zaefizadeh Data curation, Formal analysis, 5
Fatemeh Masoumi Methodology, 1
Alireza Zomorodipour Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing, 1, * 
Author information:
1Department of Molecular Medicine, Institute of Medical Genetics, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
2Department of Stem Cell and Regenerative Medicine, Institute of Medical Genetics, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
3Department of Medical Genetics, Institute of Medical Genetics, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
4Hazrat-e Rasool General Hospital, Iran University of Medical Science, Tehran, Iran
5Department of Biology, School of Sciences, Ardabil branch, Islamic Azad University, Ardabil, Iran
Abstract
Purpose:
Calumenin (CALU) is a calcium-binding protein involved in several physiological processes, exhibiting tumor-specific expression variation and emerging as a potential player in cancer progression. This study aimed to investigate the correlation between CALU and clinicopathological features in breast cancer (BC) and perform a functional assessment of CALU based on a microRNA-mediated knockdown approach.
Methods:
The BC tissues’ CALU expression was measured by q-RT-PCR. We looked at correlations between changes in CALU expression and clinicopathological characteristics. We adopted a CALU knockdown approach using an artificial microRNA (amiR), expressed through an episomal vector, in BC cell lines. Epithelial to mesenchymal transition (EMT) markers were then assessed, and cell cycle, migration, proliferation, and apoptosis were analyzed.
Results:
When compared to the normal surrounding tissues, the BC tissues showed a 3.4-fold increase in CALU expression. This was significantly correlated with clinicopathological parameters such as histological grade, Ki-67 expression, TNM stage, lymph node involvement, and vascular lymph invasion. Key EMT markers, including GSC, MMP2, TIMP1, TGF1, SLUG, ZEB1, ZEB2, SNALI1, and TWIST1, were downregulated as a result of CALU knockdown, which prevented cell migration and proliferation and caused cell cycle arrest and apoptosis in the BC cell lines.
Conclusion:
The results of the amiR-mediated knockdown approach support the findings that CALU is a potential promoter of BC, as evidenced by the upregulation of CALU in BC tissues and its correlation with clinicopathological features, which highlights its role in BC progression.
Keywords: Apoptosis, Artificial microRNA, Breast cancer, Calumenin, Cell proliferation, Metastasis
Copyright and License Information
© 2025 The Author (s).
This is an Open Access article distributed under the terms of the Creative Commons Attribution (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, as long as the original authors and source are cited. No permission is required from the authors or the publishers.
Funding Statement
This work was financially supported by Grants No. 519 and 614, awarded to A. Z. obtained from the National Institute of Genetic Engineering and Biotechnology (NIGEB), Iran.
Introduction
Calumenin (CALU) is a multiple EF-hand calcium (Ca)-binding protein and a member of the Cab45/reticulocalbin/ERC-45/CALU (CREC) protein family. The mammalian CREC family members, mainly located in various secretory pathway compartments, are characterized by multiple EF-hands and participate in different physiological processes, and Ca homeostasis is the primary outcome of their functions.1,2
Cancer is a complex and multifaceted disease characterized by uncontrolled cell growth, evasion of apoptosis, invasion of surrounding tissues, and metastasis to distant organs.3 It is imperative to comprehend the molecular mechanisms that propel cancer progression to design potent and efficacious treatment.4 Emerging evidence in recent years suggests that CALU may play a significant role in cancer promotion, and its association with more malignant phenotypes and shorter survival rates for patients has been widely reported.5,6
Metastasis is a primary cause of the majority of cancermorbidities and mortalities, and the development of effective treatment programs requires a better understanding of the molecular mechanisms behind this process.7 CALU has been suggested to promote metastasis of cancer cells by interacting with extracellular matrix (ECM) components, such as fibronectin and laminin.8 CALU also has emerged as a regulator of epithelial to mesenchymal transition (EMT), promoting the transition from an epithelial to a mesenchymal phenotype. This Ca-binding protein enhances the expression of EMT-inducing transcription factors, such as Snail, Twist, and ZEB1, while downregulating epithelial markers like E-cadherin and promoting cancer cell migration, invasion, and resistance to therapy.5,6 Furthermore, CALU may inhibit angiogenesis, a critical process for tumor growth and metastasis through fibulin-1.9,10
The human CALU gene produces 15 isoforms, among them CALU 1–14, with N-terminal signal peptides, are either localized in secretory pathway compartments or secreted into the extracellular space, and CALU-15 shuttles between nucleus and cytoplasm, due to lack of signal peptide.8 Among the CALU isoforms, CALU-15 is reportedly involved in cell migration by promoting the formation of filopodia, an actin-rich finger-like membrane protrusion with a key role in metastasis.8 CALU-15, a nuclear-localized and phosphorylation-dependent CALU isoform, promotes filopodia formation and cell migration by upregulating growth differentiation factor-15 (GDF-15), a transforming growth factor-beta (TGF-b)-like cytokine.8 GDF-15 has been shown to have both antitumor and antiproliferative properties, although this remains a highly controversial topic.11-14 Due to its pleiotropic effects, GDF-15 operates under the tight control of many regulatory pathways and participates in a variety of cellular processes.15-20 Consequently, CALU-15 was suggested as a new regulator of GDF-15.8
In contrast, the extracellular CALU isoforms suppressthe signal-regulated kinase 1 and 2 (ERK1/2) signaling and inhibit cell migration.21 These pieces of evidence suggest the potential prognostic and therapeutic values of CALU isoforms in cancers. In our previous study, we introduced a panel of CALU/ARUKA/MCM2 genes with high accuracy in discriminating the biopsies of colon and lung cancers from healthy samples, where the high expression levels of these genes are negatively correlated with the patient’s survival rate.22
The widespread involvement of microRNAs (miRNA) in many human diseases, including various cancers, makes them attractive tools for targeted therapeutic strategies.23 In this regard, synthetic miRNAs, by silencing the gene(s) of interest, have shown great promise as a new class of targeted therapeutic agents for treating various human diseases, including cancer.24 The most attractive aspect of miRNA therapeutics is its ability to target almost any gene that may not be possible with small molecules or protein-based drugs, although there are still challenges regarding their application at the clinical level.25
Although there is a wealth of evidence supporting the critical role of CALU in many cellular and cancer-related processes, little is known about its function in breast cancer (BC).26 In this study, we aimed to investigate the possible relationship between CALU expression patterns and clinicopathological and demographic characteristics among women with BC. Besides, using a CALU-specific artificial miRNA (amiR), we have adopted a miRNA-mediated knockdown strategy in two BC cell lines to address the CALU function in BC and its association with proliferation, apoptosis, and metastasis potential.
Materials and Methods
Human samples collection and cell culture
The National Institute of Genetic Engineering and Biotechnology (NIGEB) of Iran’s ethics committee accepted this work under ethics approval codes IR.NIGEB.EC.1402.11.29.D and IR.NIGEB.EC.1402.11.29.E. This study included 55 BC women who had received BC surgery at Khatam-Ol-Anbia Hospital in Tehran, Iran. Fresh tissue specimens (tumor tissues and their normal adjacent tissues) were collected in separate sterile tubes, frozen, and stored at -70°C. An expert pathologist confirmed a histologic diagnosis for all samples. Informed written consent was obtained from patients for all tissue samples by the clinicians.
The SK-BR-3 and MDA-MB-231 BC cell lines were purchased from the Iranian Biological Resource Center (Tehran, Iran) and cultured in high-glucose DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, USA), 100 U. mL-1 penicillin, and 100 μg. mL-1 streptomycin (Sigma, USA) at 37 °C in a 5% CO2-humidified incubator. After the BC cell lines had reached a 70%-80% confluent monolayer, they were passaged and used for subsequent experiments.
The cloning processes were carried out using the E. Coli strain DH5α (Stratagene, USA). For PCR, PCR cloning, and sub-clonings, MWG-Germany’s produced primers were employed (Table 1). Thermo Fisher Scientific, USA, provided the enzymes XbaI, XhoI, HindIII, StuI, PstI, T4 DNA ligase, DNase, and reverse transcriptase (M-MuLV).
Table 1.
Primers used in this study
|
Gene name
|
Accession
|
Forward primer Seq. (5’-3’)
|
Forward primer location
|
Reverse primer Seq. (5’-3’)
|
Reverse primer location
|
PCR product size (bp)
|
| ACTB |
NM_001101.5 |
GAGACCTTCAACACCCCAGCC |
Exon 4 |
AGACGCAGGATGGCATGGG |
Exon 4 |
161 |
| CALU |
NM_001219.5 |
GATGGTTAGAGATGAGCGGAG |
Exon 4 |
ATCTTCCATTGTTTCCTGTACTACT |
Exons 4-5 boundary |
142 |
| SNAI1 |
NM_005985.4 |
ATGCACATCCGAAGCCACAC |
Exon 2 |
CACTGGTACTTCTTGACATCTG |
Exon 3 |
203 |
| SNAI2 |
NM_003068.5 |
GCGATGCCCAGTCTAGAAAAT |
Exon 2 |
ACCTGTCTGCAAATGCTCTGTT |
Exon 3 |
225 |
| TIMP1 |
NM_003254.3 |
TCCTGTTGTTGCTGTGGCTGA |
Exon 2 |
CCTTTATACATCTTGGTCATCTTG |
Exons 3-4 boundary |
181 |
| MMP2 |
NM_004530.6 |
TTGGCTACACACCTGATCTGG |
Exon 3 |
GAGTCCGTCCTTACCGTCAAA |
Exon 4 |
185 |
| GSC |
NM_173849.3 |
GAAAGTGGAGGTCTGGTTTAA |
Exons 2-3 boundary |
TACCTTCCTCTTCCCTCTTCT |
Exon 3 |
143 |
| TWIST1 |
NM_000474.4 |
TACATCGACTTCCTCTACCAG |
Exon 1 |
GGAAACAATGACATCTAGGTCTC |
Exons 1-2 boundary |
203 |
| ZEB1 |
NM_001128128.3 |
CAGATGATGAAGACAAACTGCATA |
Exon 2 |
CCCTTCCTTTCCTGTGTCAT |
Exons 2-3 boundary |
176 |
| ZEB2 |
NM_014795.4 |
GGACAGATCAGCACCAAATG |
Exons 6-7 boundary |
ATGTGCGAACTGTAGGAACCA |
Exon 8 |
190 |
| TGFB1 |
NM_000660.7 |
CTATTGCTTCAGCTCCACGGA |
Exons 5-6 boundary |
AGGACCTTGCTGTACTGCGT |
Exons 6-7 boundary |
171 |
| GDF-15 |
NM_004864.4 |
AGAAGTGCGGCTGGGATCC |
Exons 1-2 boundary |
TCTTGCAAGGCTGAGCTGAC |
Exon 2 |
180 |
| GAPDH |
NM_001289746.2 |
AAGGTGAAGGTCGGAGTCAAC |
Exon 2 |
GGGGTCATTGATGGCAACAATA |
Exon 3 |
102 |
Plasmids
Construction of the CALU-specific amiR-6 was described previously.27 In summary, sequence comparison of fifteen CALU transcript isoforms led to the identification of a conserved sequence (GGAAACAATGGAAGATATA) that served as a template for the design of amiR-6 capable of targeting both nuclear and non-nuclear CALU variants.
A previously constructed CALU-specific amiR expressing plasmid, namely FIX-Int-miR6, expressing amiR-6,27 was modified by replacing the FIX cDNA with a 747 bp EGFP-coding DNA to end up with plasmid EGFP-Int-miRNA-6 (amiR-6). The forward and reverse primers used for PCR amplification of EGFP were designed to contain a mammalian Kozak sequence (GCCRCCATGG)28 in the start codon context and a TAA codon as a stop codon, which ensured higher translational efficiency. In the constructed plasmid (EGFP-Int-miRNA-6), the CALU-specific amiR-6 coding sequence was inserted in the EGFP 5’-UTR (Figure 1). A miRNA-less EGFP expressing plasmid was constructed and used as a negative control (mock). Restriction analysis and nucleotide sequence analysis were used to verify the recombinant plasmids. A comparison of nucleotide sequences against the gene bank was done using BLAST.29
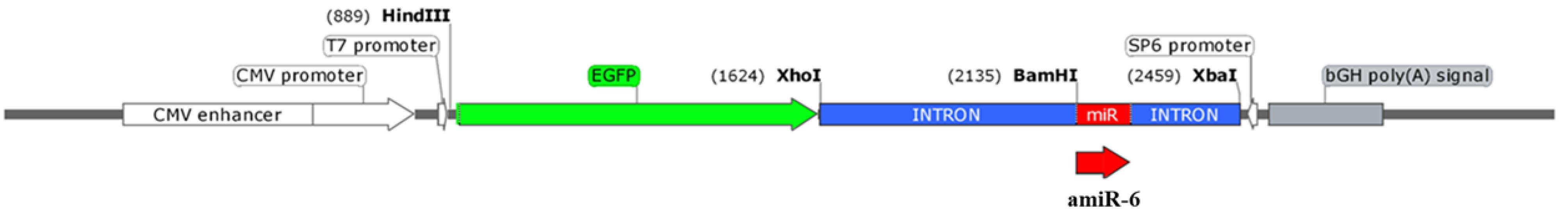
Figure 1.
A schematic view of EGFP-Int-amiR-6 expression cassette, inserted within the pcDNA3 plasmid. The CALU-specific miRNA (amiR-6, Red), within a truncated FIX intron 1 (Blue), downstream to an EGFP coding sequence (Green) are shown
.
A schematic view of EGFP-Int-amiR-6 expression cassette, inserted within the pcDNA3 plasmid. The CALU-specific miRNA (amiR-6, Red), within a truncated FIX intron 1 (Blue), downstream to an EGFP coding sequence (Green) are shown
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from tissues and cells using a High Pure RNA Isolation Kit (Roche, Germany). A NanoDrop spectrophotometer was used to measure RNA concentrations (Thermo Fisher, USA). PrimeScriptTM 1st Strand cDNA Synthesis Kit (Takara Bio, Japan) was applied to perform reverse transcription on cDNA. The concentration and purity of the extracted cDNA were estimated using a NanoDrop Spectrophotometer (Thermo Scientific, USA). PCR was used to assess the quality of the generated cDNA using GAPDH-specific primers. The levels of expressed amiR-6 and mRNAs were compared to the levels of endogenous U6 and GAPDH, respectively, to normalize their expression. The 2-ΔΔCt method was used for evaluating the real-time PCR results.30 The assays were carried out in triplicates.
Cell transfection
The BC cell lines, plated in individual wells of 6-well plates, underwent transfection with amiR-6 or mock through Lipofectamine 2000 (Invitrogen), as per the manufacturer’s guidelines. After 48h, the cells were harvested, and the later assays were performed.
Cell proliferation assay
The amiR-6 effect on the proliferation of BC cell lines was evaluated through a trypan blue dye viability assay or an exclusion test. Before the trypan blue exclusion test, a 24-well plate was seeded with 8 × 104 SK-BR-3 or MDA-MB-231 cells. Afterward, the cells underwent a transfection process involving the introduction of either amiR-6 or mock plasmid. Following 48 hours post-transfection, the number of living BC cells was determined through hemocytometer counting.
Cell cycle distribution analysis
For the cell cycle analysis of the transfected BC cell lines after 48 hours, a total of 1 × 106 cells per group were incubated with RNase A (10 mg. mL-1) for 1 h at 37 °C and stained with propidium iodide (PI) (50 µg. mL-1) in dark. The stained cells were analyzed with a BD FACSCalibur flow cytometer and FlowJo software (version 7.6.1). To ensure reproducibility, the experiment was repeated four times.
Evaluation of apoptosis
Cell apoptosis was detected by flow cytometry analysis.31 Briefly, the BC cell lines were seeded onto 6-well plates and transfected with amiR-6, or mock. The transfected BC cell lines were rinsed with phosphate-buffered saline (PBS) and re-suspended in 100 µL binding buffer at 48 h post-transfection. A two-step staining was performed on the cells, once with 5 µL of Annexin V-FITC (1 µg.mL-1) for 10 minutes and once with 5 µL of PI (1 µg.mL-1) for 5 minutes at room temperature, and in the dark. In the last step, FlowJo software (version 7.6.1) and a BD FACSCalibur flow cytometer were used to evaluate the apoptotic cells.
Acridine orange/ethidium bromide (AO/EB) staining was used to detect apoptotic cells. The BC cells were incubated in 6 well plates up to 80% confluency before transfection with amiR-6 or mock and incubation at 37 °C for 48 h. The cells were washed twice with PBS before being exposed to a 1:1 combination of acridine orange and ethidium bromide (Sigma-Aldrich, USA) solution (100 µg.mL-1 each). Using an inverted fluorescent microscope, the cells were instantly analyzed.
Wound healing assay
To perform a wound-healing assay, the transfected cells were grown in four-well tissue culture plates to 80% confluency. Using a sterile pipette tip, a gentle scrape was made to create a small linear scratch in the confluent monolayer before being washed with PBS twice to remove the detached cells and incubated in a low serum medium (1% FBS) for 72 h at 37 °C in 5% CO2 and imaged every 24 h, using an inverted microscope and analyzed by image analysis software (ImageJ, National Institute of Health, Bethesda, MD, USA). The wound healing degree was estimated by the distance passed by cells into the scratched area. The presented data is an average of three independent experiments.
Transwell migration assay
To investigate the amiR-6 impact on migration, a transwell assay was carried out on the transfected BC cells using transwell plates (SPL) with an 8.0 µm pore size. Briefly, 1 × 105 MDA-MB-231 and SK-BR-3 cells were transfected with amiR-6. After 48 h, the cells were trypsinized, rinsed with PBS, added to a serum-free medium, and cultured in the upper chamber of the inserts. As a chemoattractant, 10% FBS medium was added to the lower chamber. After culturing with 5% CO2 at 37 °C for 48 h, cotton wool was used to gently remove the cells that did not migrate through the pores. The inserts were then observed by inverted microscopy after being fixed with 20% methanol, stained with 0.2% crystal violet, and dried. Five distinct fields were used to count the migrated cells.
Statistical analysis
The normal distribution of data was confirmed using SPSS version 22 software and the Kolmogorov-Smirnov test.32 The relative gene expression was obtained using the Pfaffl method.33 To investigate the possible relationship between the CALU transcription and clinicopathological features, SPSS version 22 software, independent-sample t-test, one-way ANOVA, and Duncan’s multiple range test were used. A 95% confidence interval was considered for all quantitative tests, and P values less than 0.05 were considered significant.
Results and Discussions
Evaluation of relative CALU mRNA expression in tumor vs. normal breast tissues
Evaluation of CALU expression among BC patients, based on the results obtained from RT-qPCR, showed a 3.4-fold increase in CALU transcript levels in breast tumor tissues compared to normal adjacent tissues (Figure 2A). Our data aligns with findings from Pan-cancer analyses, which reveal that CALU is consistently upregulated in BC, particularly the aggressive basal-like (triple-negative) subtype, and associated with poor prognoses.26 In vitro studies further demonstrate high CALU expression in triple-negative BC (TNBC) reverses EMT and inhibits migration, suggesting its significant role in promoting the mesenchymal transition and aggressive potential of this subtype.26 While the specific mechanisms and relationship to hormone receptor status warrant further investigation, CALU holds promise as a diagnostic or prognostic marker, especially in TNBC. Further investigation of CALU expression in Luminal A/B and HER2-enriched subtypes35 could also reveal valuable insights into its role in BC progression.
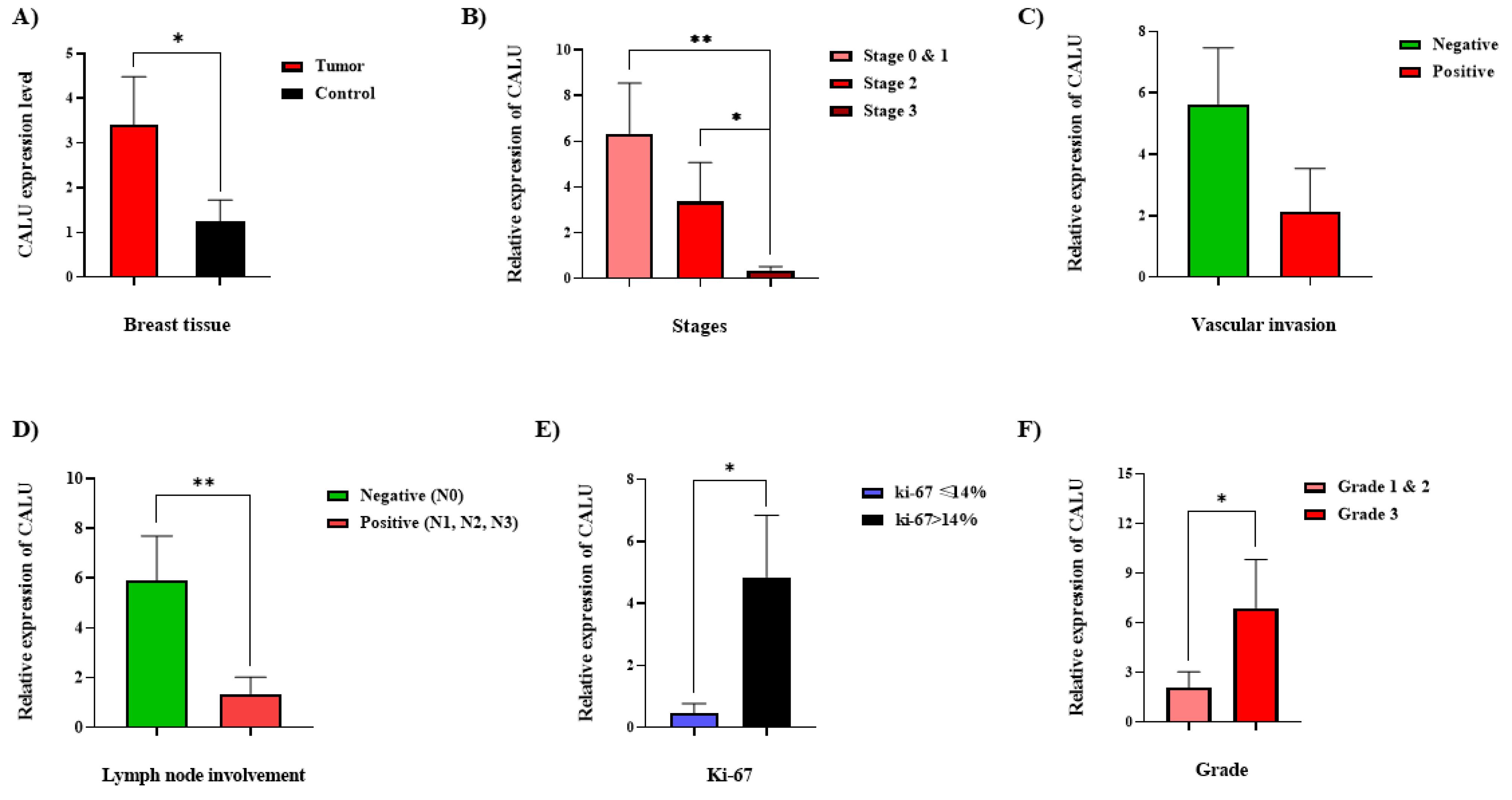
Figure 2.
The CALU transcription assessment, concerning clinicopathological features of the BC patients. (A) Comparison of the CALU transcription in tumor and normal tissues in women with BC. The CALU transcriptions at different TNM stages of BC (B), in breast tumor samples with the negative status of vascular invasion compared with positive status (C), in tumor samples from patients with negative status of lymph node involvement compared with those of samples with positive status (D), at different levels of ki-67 (E), and different histological grades among BC tissues (F). The use of asterisks denotes notable variances in comparison to the control group. * = P ≤ 0.05, ** = P ≤ 0.01
.
The CALU transcription assessment, concerning clinicopathological features of the BC patients. (A) Comparison of the CALU transcription in tumor and normal tissues in women with BC. The CALU transcriptions at different TNM stages of BC (B), in breast tumor samples with the negative status of vascular invasion compared with positive status (C), in tumor samples from patients with negative status of lymph node involvement compared with those of samples with positive status (D), at different levels of ki-67 (E), and different histological grades among BC tissues (F). The use of asterisks denotes notable variances in comparison to the control group. * = P ≤ 0.05, ** = P ≤ 0.01
Other studies also revealed a 10-fold increase in CALU expression in breast tumor interstitial fluid,34 and a 4-fold increase in CALU expression in a metastatic breast cell line compared to non-metastatic cells.35 Variation in the CALU expression during tumor progression was reported in other cancers, although the variation pattern is tissue-specific.1 In most cancer types, such as oral cancer,36 colon and lung cancers,22 and more malignant gliomas,5 enhancement of the CALU expression has been evidenced. In contrast, down-regulation of CALU in metastatic cell lines of head and neck,37,38 hepatocellular and pancreatic carcinoma,2,21 and lung squamous cell carcinoma,39 were shown. Differential regulation of the CALU has been observed in various malignancies, indicating its potential as a prognostic marker.40
Correlations between clinicopathological features and CALU expression
Classification of the BC tissue samples was performed based on various clinicopathological features such as pathological type, histological grade, Ki-67 status, tumor size, lymph node involvement, TNM (stands for tumor size, lymph node number and position, and metastasis) staging, receptor status, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2), in addition to features such as necrosis, calcification, vascular invasion, and perineural invasion, outlined in Table 2. The analysis of the CALU expression variations, considering the clinicopathological features, demonstrated significant correlations between the CALU expression and features, including TNM staging, lymph node involvement, vascular invasion, histological grading, and Ki-67 index (Figure 2).
Table 2.
Correlations between the BC clinicopathological features and CALU expression.
|
Clinicopathological features
|
Different states
|
The number of patients (%)
|
CALU expression (mean±SEM)
|
P
value
|
| Age (year) |
˂45 |
15 (27.27) |
3.28 ± 1.59 |
0.79 |
| 45-55 |
22 (40) |
4.56 ± 2.18 |
| ˃55 |
16 (29.09) |
2.34 ± 1.49 |
| Not known |
2 (3.63) |
NA |
| Pathological type |
IDC |
35 (63.63) |
3.67 ± 1.39 |
0.82 |
| ILC |
9 (16.36) |
3.74 ± 2.81 |
| Others |
8 (14.54) |
3.11 ± 2.73 |
| Not known |
3 (5.45) |
NA |
| Histological grade |
1 & 2 |
37 (67.27) |
2.13 ± 0.90 |
0.03 |
| 3 |
16 (29.09) |
6.76 ± 2.89 |
| Not known |
2 (3.63) |
NA |
| Ki-67 |
≤ 14% |
7 (12.72) |
0.55 ± 0.41 |
0.05 |
| ˃14% |
26 (47.27) |
4.85 ± 1.99 |
| Not known |
22 (40) |
NA |
| Tumor size |
≤ 2 |
22 (40) |
4.76 ± 1.80 |
0.39 |
| 2-5 |
28 (50.90) |
2.86 ± 1.52 |
| ˃ 5 |
2 (3.63) |
0.99 ± 0.80 |
| Not known |
3 (5.45) |
NA |
| Lymph node Involvement |
Negative |
26 (47.27) |
5.80 ± 2.02 |
0.005 |
| Positive |
27 (49.09) |
1.34 ± 0.77 |
| Not known |
2 (3.63) |
NA |
| TNM stage |
0 & 1 |
16 (29.09) |
6.37 ± 2.36 |
0.02 |
| 2 |
24 (43.63) |
3.37 ± 1.76 |
| 3 |
12 (21.81) |
0.34 ± 0.16 |
| Not known |
3 (5.45) |
NA |
| ER |
Positive |
41 (74.54) |
4.34 ± 1.40 |
0.56 |
| Negative |
11 (20) |
0.81 ± 0.32 |
| Not known |
3 (5.45) |
NA |
| PR |
Positive |
39 (70.90) |
3.92 ± 1.39 |
0.83 |
| Negative |
13 (23.63) |
2.60 ± 1.68 |
| Not known |
3 (5.45) |
NA |
| HER-2 |
Positive |
12 (21.81) |
4.10 ± 2.36 |
0.99 |
| Negative |
26 (47.27) |
2.02 ± 0.97 |
| Not known |
17 (30.90) |
NA |
| Necrosis |
Present |
25 (45.45) |
3.25 ± 1.69 |
0.62 |
| Absent |
21 (38.18) |
3.79 ± 1.68 |
| Not known |
9 (16.36) |
NA |
| Calcification |
Present |
9 (16.36) |
2.97 ± 2.20 |
0.84 |
| Absent |
38 (69.09) |
4.20 ± 1.44 |
| Not known |
8 (14.54) |
NA |
| Vascular invasion |
Present |
27 (49.09) |
2.12 ± 1.42 |
0.01 |
| Absent |
23 (41.81) |
5.64 ± 1.84 |
| Not known |
5 (9.09) |
NA |
| Perineurial invasion |
Present |
11 (20) |
3.11 ± 2.32 |
0.60 |
| Absent |
34 (61.81) |
3.72 ± 1.43 |
| Not known |
10 (18.18) |
NA |
The samples were categorized into groups 1, 2, and 3, which correspond to stages 1/0, 2, and 3, respectively, based on the TNM staging (Figure 2B). According to an intergroup analysis, samples from stages 1.0 and 3 had the highest and lowest CALU transcription levels, respectively, with a significant difference (P-value < 0.02). Patients were classified as either positive or negative for vascular invasion depending on whether the disease recurred or did not. Patients with negative vascular invasion had higher levels of CALU expression than those with positive vascular invasion; however, this difference was not statistically significant (P value = 0.052) (Figure 2C). Patients’ status was classified as either positive or negative based on the involvement of lymph nodes. Compared to individuals with positive status, those with negative status had noticeably greater amounts of CALU transcripts (Figure 2D).
The analyses revealed that in BC tissues, the highest amount of CALU transcription happened between stages 0 and 1 when vascular invasion and lymph node involvement were still absent. This finding indicates that, while still higher than in normal tissues, CALU expression gradually declines as BC advances and lymph nodes and vascular invasion occur. Afterward, the transcription level of CALU decreased as the number of affected lymph nodes increased. The tumor microenvironment (TME) may be the reason why CALU expression levels rise during the beginning of BC and fall subsequently. Even though CALU expression has decreased in cancerous tissue, it is still greater than in healthy tissues.
To distinguish between the luminal A and luminal B stages of BC, a midpoint of 14% was proposed as the ideal limit for Ki-67 evaluation.41 Our analysis showed that samples with ki-67 > 14% had higher CALU mRNA expression than samples with ki-67 < 14% (Figure 2E).
There were notable changes in CALU expression between the two groups of investigated samples based on histological grade. Tumor samples of grades 1 and 2 were included in group 1, while only grade 3 tumor samples were included in group 2. Grade 3 samples had a greater average CALU mRNA level than grades 1 and 2 samples (Figure 2F).
It is advised to employ Histological grade42 and Ki-67,43 two significant prognostic factors in the early stages of BC, together for a more precise evaluation.7 Both of these characteristics and the CALU transcription were directly correlated in our study, confirming the link between the CALU and the invasive nature of BC.5,8
Evaluation of the effects of CALU knockdown on the BC cells
When CALU transcription was evaluated in SK-BR-3 cells transfected with a plasmid encoding amiR-6, CALU was successfully knocked down (Figure 3A). GDF-15 expression was likewise reduced in these cells as a result of amiR-6-mediated CALU knockdown (Figure 3B). According to a previous study, the colorectal SW480 cell line’s GDF-15 is stimulated by the CALU nuclear isoform,CALU-15, which increases cell motility and filopodia production in colon cancer.8 Numerous biological processes have been reported as being regulated by GDF-15, which exhibits both tumor promoter and tumor suppressor properties.44,45

Figure 3.
The expression levels ofCALU (A), GDF-15 (B), and EMT markers (C) in the SK-BR-3 cell line, using qRT-PCR, following the AmiR-6 overexpression, at 48h post-transfection. The names of markers are given. * = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001
.
The expression levels ofCALU (A), GDF-15 (B), and EMT markers (C) in the SK-BR-3 cell line, using qRT-PCR, following the AmiR-6 overexpression, at 48h post-transfection. The names of markers are given. * = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001
GDF-15 promotes the migratory and invasive phenotype of BC cells in vitro by activating the actin cytoskeleton-regulating genes alpha Parvin (PARVA), RhoA, Rho-associated protein kinase-1 (ROCK-1), and Facsin-1, according to a systematic review conducted to revisit the potential involvement of GDF-15 in cell metastasis in various cancer types.46 It has previously been suggested that GDF-15 contributes to the development of BC by triggering signaling pathways that regulate EMT and cellular invasion,47 as well as the acquisition of characteristics similar to those of cancer stem cells.48,49
While GDF-15 promotes BC cells’ migratory and invasive characteristics, its effects vary depending on the cancer type. For instance, GDF-15 has been demonstrated to have proapoptotic and anti-metastatic properties in colon carcinoma cells,50 lung cancer,51 and ovarian and prostate cancer cell lines.52 There is also evidence that overexpression of GDF-15 inhibits the tumorigenicity of LN-Z308 glioblastoma cell line.53
Apart from its carcinogenic activity, CALU’s isoforms found in the ECM and secretory pathway were also found to have a tumor-suppressive function.21,54 The interaction between the extracellular CALU isoforms and fibulin-1, a member of the ECM fibulin family that is directly linked to fibronectin, is the basis for the tumor suppressor effects of the CALU isoforms.55 By establishing a compound with fibulin 1, the extracellular CALU isoforms stop MMP-13 from degrading it. This cooperation restricts cell migration via an integrin and extracellular signal control, as well as the protein kinases 1 and 2 (ERK1/2) signaling pathway.21 Through its interactions with integrins, syndecans, and cell surface receptors, Fibilin-1 mediates cell adhesion.56-58
The amiR -mediated CALU-knockdown suppressed EMT in the BC cells
Suppression of EMT markers is anticipated to reduce the invasive and metastatic potential of BC cells in vivo, as EMT is a crucial and first stage in the metastasis cascade. To break through the basement membrane, intravasate, circulate, and extravasate at distant locations, cancer cells must undergo EMT.59 It has previously been hypothesized that by reversing EMT, CALU knockdown may disrupt metastasis processes and limit the ability of cancer cells to spread.60 To examine this hypothesis, the expression assessment of EMT-related markers, including SNAI1, TGF-b1, ZEB1, ZEB2, SNAI2 (SLUG), GSC, MMP2, TIMP1, and TWIST in the SK-BR-3 BC cell line, was performed at 48 h of post-transfection with amiR6. Overexpression of amiR-6 in the SK-BR-3 BC cell line showed inhibitory effects on the studied EMT markers (Figure 3C). This result could have been caused by GDF-15 downregulation (Figure 3B), which is corroborated by earlier studies showing that GDF-15 promotes the expression of EMT markers, SNAIL and TWIST in colorectal cancer.48,61,62 These two proteins play a role in suppressing the epithelial phenotype by directly inhibiting the expression of E-cadherin.63,64 The potential of CALU as a hallmark of EMT is supported by the results acquired thus far from the CALU knockdown technique in the SK-BR-3 cell line, which are in accord with other reports.34,35,40,54,65
The regulatory role of CALU in EMT in BC was validated by Chen et al in a recent pan-cancer screening study. This investigation demonstrated that CALU, a crucial element of the TME, promotes cell migration through EMT.26 To further restrict invasion and metastasis, it appears that CALU knockdown may have an impact on the interactions between cancer cells and stromal cells in vivo.
Research on gliomas has shown that EMT phenotype is closely linked to genes relevant to CALU.5 Several immunohistochemical tests, the Gene Expression Omnibus (GEO) database, and the Cancer Genome Atlas (TCGA) have also revealed CALU to be one of the most prevalent proteins expressed in cancer-associated fibroblasts (CAF), supporting its inclusion in the signature of EMT-related genes in gastric cancer.66
According to the findings of the previous stage, which demonstrated the impact of CALU on GDF-15 expression, the results of the evaluation of the EMT marker of cells treated with amiR-6 validate the relationship between CALU and EMT markers as well as its regulatory function in cancer-related processes. The results obtained from the evaluation of the EMT marker of cells treated with amiR-6, in line with the inhibitory function of CALU on GDF-15 expression, support the association of CALU with EMT markers and cancer-related processes. While in vivo studies are needed to validate these findings, our results suggest that CALU is a promising therapeutic target for inhibiting BC metastasis.
Effect of amiR -mediated degradation of CALU on proliferation, apoptosis, and migration of BC cell lines
CALU knockdown inhibited the proliferation of BC cell lines
The SK-BR-3 cell line was assessed alongside the MDA-MB-231 cell line to examine the impact of amiR-mediated CALU knockdown on BC cell proliferation after the crucial influence of CALU on EMT was demonstrated in this cell line. Cell proliferation was then evaluated at 24 and 48 hours, using trypan blue staining techniques. At 48 hours after transfection, amiR-6 dramatically decreased cell proliferation in both treated BC cell lines, when compared to the control group (Figure 4A, B). Using cell cycle analysis, we further validated this result (Figure 4C, D). The G1 phase cell population of the amiR-6 transfected group was significantly higher than that of the control group, according to the data.

Figure 4.
The proliferation assessment of the BC cell lines, SK-BR-3 (upper row), and MDA-MB-231 (lower row), before and after the amiR-6 overexpression. (A, B) Trypan blue staining assessments. (C, D) Cell cycle analysis. * P < 0.05, ** P < 0.01, *** P < 0.001)
.
The proliferation assessment of the BC cell lines, SK-BR-3 (upper row), and MDA-MB-231 (lower row), before and after the amiR-6 overexpression. (A, B) Trypan blue staining assessments. (C, D) Cell cycle analysis. * P < 0.05, ** P < 0.01, *** P < 0.001)
Knockdown of CALU-induced apoptosis in BC cell lines
At 48 hours after amiR-6 transfection, the cell lines MDA-MB-231 and SK-BR-3 BC were simultaneously stained with ethidium bromide and acridine orange to examine the impact of CALU knockdown on apoptosis in BC cells. Acridine orange exhibits green or red fluorescences to dsDNA or ssDNA/RNA, depending on whether it is absorbed by living or dead cells. On the other hand, ethidium bromide only penetrates non-viable cells, where it attaches itself to DNA and emits red fluorescence.67
The cells transfected with amiR-6 were yellow to orange, suggesting the presence of apoptosis, whereas both BC cell lines analyzed in the control group were green, showing the lack of apoptosis (Figure 5A, C). The apoptotic cells were confirmed and measured by flow cytometry analysis. Approximately 15.44% of the transfected SK-BR-3 cells accounted for apoptotis (early and late apoptosis) (Figure 5B), while 23.21% of the transfected MD-MB-231 cells accounted for apoptotis (Figure 5D). Estimation of the percentage of apoptosis in both SK-BR-3 and MDA-MB-231 cells after transfection with amiR-6, suggests that the CALU-knockdown significantly increased apoptosis in the BC cells (Figure 5E).

Figure 5.
The impact of the AmiR-6 overexpression on apoptosis in the BC cell lines. Acridine orange/ethidium bromide (AO/EB) staining (A, B). Apoptosis analysis using PI staining (PI, Y-axis) and Annexin-FITC (X-axis) (C, D). (Q1 = necrotic cells, Anx- /PI + ; Q2 = late apoptotic cells, Anx + /PI + ; Q3 = early apoptotic cells, Anx + /PI-; and Q4 = living cells, Anx-/ PI-). E) “Quantification of apoptosis (%) in SK-BR-3, and MDA-MB-231cells after CALU knockdown”
.
The impact of the AmiR-6 overexpression on apoptosis in the BC cell lines. Acridine orange/ethidium bromide (AO/EB) staining (A, B). Apoptosis analysis using PI staining (PI, Y-axis) and Annexin-FITC (X-axis) (C, D). (Q1 = necrotic cells, Anx- /PI + ; Q2 = late apoptotic cells, Anx + /PI + ; Q3 = early apoptotic cells, Anx + /PI-; and Q4 = living cells, Anx-/ PI-). E) “Quantification of apoptosis (%) in SK-BR-3, and MDA-MB-231cells after CALU knockdown”
Knockdown of CALU inhibited the migration of BC cell lines
A transwell experiment was used to investigate the impact of amiR-6-mediated CALU-knockdown on the migration of two BC cell lines. The results demonstrated a significant decrease in the migratory capacity of both cell lines 48 hours after transfection (Figure 6). According to our results, fewer migratory cells adhered to the bottom of the chamber when CALU was knocked down by amiR-6 (Figure 6A, B). In addition to the transwell migration test, a wound healing experiment was used to show how CALU knockdown affected the migration of BC cells. The migratory rate of the BC cells in the scratched areas decreased significantly for both BC cell lines after amiR-6 expression (Figure 6C, D). The conclusion, that CALU can successfully affect BC cell migration, was corroborated by the results of the transwell migration and wound healing assays.
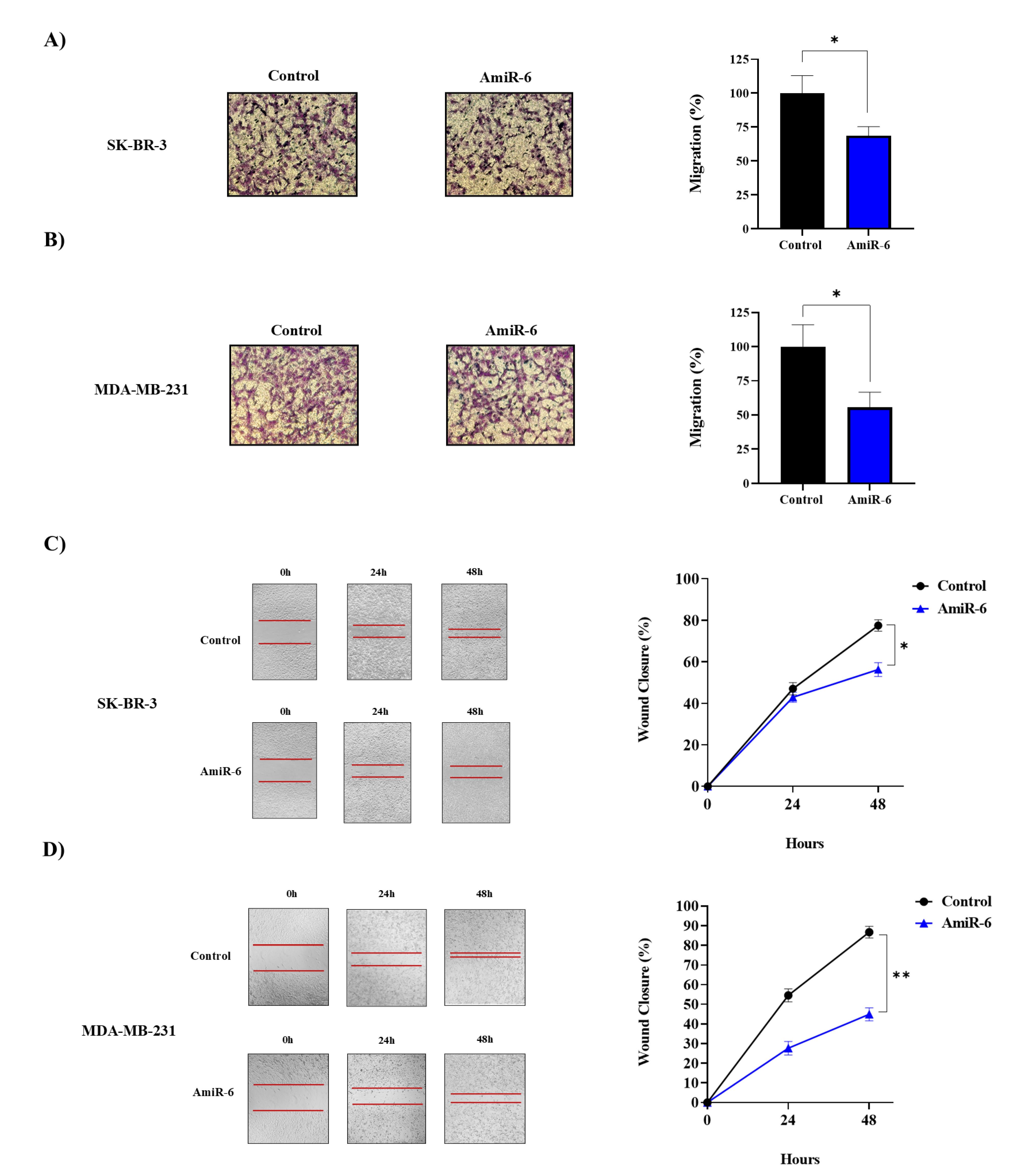
Figure 6.
The effect of the amiR6-mediated CALU knockdown on the migration rates of the BC cell lines, by performing wound healing assay. The microscopic picture of the cells (A), and the migration rate (%) assessment of the cells at 0, 24, and, 48 h of post-transfection (B). The distances of the cell-free surface, are measured by image j software (C, D). * = P ≤ 0.05, ** = P ≤ 0.01.
.
The effect of the amiR6-mediated CALU knockdown on the migration rates of the BC cell lines, by performing wound healing assay. The microscopic picture of the cells (A), and the migration rate (%) assessment of the cells at 0, 24, and, 48 h of post-transfection (B). The distances of the cell-free surface, are measured by image j software (C, D). * = P ≤ 0.05, ** = P ≤ 0.01.
After demonstrating the beneficial effect of CALU on EMT in SKBR3, we demonstrated that CALU knockdown results in cell cycle arrest, a higher percentage of necrotic and apoptotic cells, and decreased proliferation and migration in both BC cell lines by using the knockdown technique in both SKBR3 and MDA-MB-231 cell lines. These findings are consistent with the EMT marker analysis results, demonstrating the critical function of CALU in BC metastatic promotion. In colon and lung tumors, our group recently described the involvement of CALU in inducing metastases.22 These results are in line with reports that indicate CALU plays a role in increasing the invasion, migration, metastasis, and proliferation of cancer cells during the carcinogenesis process.68,69 In more recent work, Li and colleagues reported the promoting function of CALU in lung adenocarcinoma progression by enhancing cell proliferation, migration, and invasion.40 Consistent with these findings, upregulation of CALU and other known CAF markers, such as periostin (POSTN, or PSTN), α-smooth muscle actin, and podoplanin, was demonstrated in lung adenocarcinoma, suggesting CALU, as a CAF-derived molecule, act effectively in cancer promotion.9 Focusing on TNBC-specific mechanisms, that allow the expansion and activity of self-renewing therapy-resistant cancer stem cells (CSC), CALU was found among the Top 30 upregulated genes in CD44-high/CD24-low cluster and suggested as a CSC regulator in TNBC and other cancers.70 Thus, strategies aimed at inhibiting CALU expression or activity could potentially disrupt cancer progression, sensitize cancer cells to apoptosis, inhibit EMT, and reduce metastasis.71
CALU regulates numerous cellular functions in significant ways.72,73 It promotes vascular calcification by reducing gamma-carboxylation and inhibiting the activation of matrix Gla protein.57,58 Moreover, CALU is crucial for maintaining calcium homeostasis,1 and controlling the uptake of Ca2 + during smooth muscle and cardiac contraction and excitation.74 Furthermore, CALU plays a part in iron-dependent cell death, gene alterations, and TME remodeling.6 It has been proposed that CALU is an upstream regulator that affects several pathways and, consequently, the development of cancer.40 Both in vitro and in vivo, the amiR-mediated CALU knockdown of mucosal melanoma cells caused apoptosis and cell cycle arrest while suppressing cell growth, migration, invasion, and metastasis, most likely by blocking phosphorylated extracellular signal-regulated kinase (ERK) signaling.69 Additionally, it has been demonstrated that CALU regulates apoptosis, and that apoptosis dysregulation leads to cancer cell survival and chemotherapy resistance.68,75,76
The CALU knockdown’s impact on cell migration may be mediated through disruptions in key signaling pathways such as integrin, MAPK, and PI3K/AKT. Integrins, essential for cell-ECM interactions and migration, could be affected by CALU’s role in EMT and TME modulation.77,78 Furthermore, CALU may influence the MAPK pathway via GDF-15,79,80 and GDF-15 can activate AKT, impacting downstream targets involved in cell survival, growth, and migration.81,82 However, further investigation is required to fully elucidate the specific mechanisms and confirm the involvement of these pathways in CALU’s regulation of BC cell migration.
In tumor tissues from mice with BC, Menon, and colleagues found that the CALU-1 isoform was up-regulated whereas CALU-2 expression remained unchanged,83 supporting the idea that CALU isoforms are differentially expressed. In this context, it is noteworthy that CALU-15 has been shown to have the opposite effect on tumor progression when compared to other CALU isoforms of the secretory pathway.8,21 This finding may contribute to the regulating role of CALU in processes connected to cancer. Given the proposed function of CALU-15, more research is necessary to clarify the mechanisms of action of CALU isoforms and their potential connection to processes connected to BC.
The CALU-mediated knockdown approach with CALU-specific miRNA adopted in this study revealed the impact of CALU on the gene(s) or pathways associated with BC progression. However, when it comes to cancer treatment, this approach could also provide a suitable strategy for targeted therapy approaches by silencing gene(s) that are involved in tumorigenesis. The miRNA-mediated knockdown holds promise, because of its ability to modulate gene expression levels, without turning it off, impacting key pathways involved in cancer progression. The clinical implications of this approach are vast, from direct tumor suppression to combat therapeutic resistance, even in personalized cancer therapies, and in combination cancer therapies.23,84,85
Our study has limitations, including its in vitro nature, limited cell lines, potential off-target microRNA effects, unclear molecular mechanisms, and small clinical sample size. These factors should be considered when interpreting the results. Future research should involve in vivo studies, more diverse cell lines, stricter microRNA controls, mechanistic investigations, larger clinical cohorts, and blind reviewing of clinicopathological features.
Conclusion
Altogether, the evidence presented in this study, including the upregulation of CALU in BC tissues and its correlation with the main clinicopathological features in BC patients, in addition to the result of CALU knockdown, have confirmed the metastasis-promoting role of CALU, which occurs via inhibiting apoptosis and enhancing the proliferation, migratory and invasive abilities of BC cells. These intriguing observations warrant further investigation into the specific mechanisms of CALU’s regulatory role. Understanding these mechanisms and their association with different CALU isoforms could lead to the development of CALU-targeted therapeutic interventions tailored to specific types of cancer, thus offering a promising path for clinical translation. In addition, our study provides valuable experimental and foundational data for further research and a more comprehensive understanding of the molecular function of CALU in BC.
A key strength is our use of a amiR-mediated knockdown approach, which may offer advantages in terms of long-term gene silencing compared to traditional siRNA methods. Furthermore, our study provides strong evidence for a correlation between CALU expression and a comprehensive set of clinicopathological features in BC patients. We also performed a detailed analysis of the effects of CALU knockdown on various cellular processes. However, like many in vitro studies, our findings may not fully recapitulate the complexity of the TME in vivo. In addition, the precise molecular mechanisms underlying the effects of CALU knockdown remain unclear, and our clinical sample size was relatively small. Future studies may address these limitations.
Competing Interests
None
Data Availability Statement
The data that supports the findings of this study are available in the supporting materials of this article. Further detailed experimental data that support the findings of this study are available from the corresponding author upon reasonable request.
Patient Consent Statement
Concerning the tissue samples, used in this work, written informed consent was obtained for all cases by clinicians.
References
- Honoré B, Vorum H. The CREC family, a novel family of multiple EF-hand, low-affinity Ca2 + -binding proteins localised to the secretory pathway of mammalian cells. FEBS Lett 2000; 466(1):11-8. doi: 10.1016/s0014-5793(99)01780-9 [Crossref] [ Google Scholar]
- Xu S, Xu Y, Chen L, Fang Q, Song S, Chen J. RCN1 suppresses ER stress-induced apoptosis via calcium homeostasis and PERK-CHOP signaling. Oncogenesis 2017; 6(3):e304. doi: 10.1038/oncsis.2017.6 [Crossref] [ Google Scholar]
- Seyfried TN, Huysentruyt LC. On the origin of cancer metastasis. Crit Rev Oncog 2013; 18(1-2):43-73. doi: 10.1615/critrevoncog.v18.i1-2.40 [Crossref] [ Google Scholar]
- Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol 2022; 15(1):129. doi: 10.1186/s13045-022-01347-8 [Crossref] [ Google Scholar]
- Yang Y, Wang J, Xu S, Shi F, Shan A. Calumenin contributes to epithelial-mesenchymal transition and predicts poor survival in glioma. TranslNeurosci 2021; 12(1):67-75. doi: 10.1515/tnsci-2021-0004 [Crossref] [ Google Scholar]
- Du Y, Miao W, Jiang X, Cao J, Wang B, Wang Y. The epithelial to mesenchymal transition related gene calumenin is an adverse prognostic factor of bladder cancer correlated with tumor microenvironment remodeling, gene mutation, and ferroptosis. Front Oncol 2021; 11:683951. doi: 10.3389/fonc.2021.683951 [Crossref] [ Google Scholar]
- Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol 2020; 60:14-27. doi: 10.1016/j.semcancer.2019.08.012 [Crossref] [ Google Scholar]
- Feng H, Chen L, Wang Q, Shen B, Liu L, Zheng P. Calumenin-15 facilitates filopodia formation by promoting TGF-β superfamily cytokine GDF-15 transcription. Cell Death Dis 2013; 4(10):e870. doi: 10.1038/cddis.2013.403 [Crossref] [ Google Scholar]
- Kunita A, Morita S, Irisa TU, Goto A, Niki T, Takai D. MicroRNA-21 in cancer-associated fibroblasts supports lung adenocarcinoma progression. Sci Rep 2018; 8(1):8838. doi: 10.1038/s41598-018-27128-3 [Crossref] [ Google Scholar]
- Xie L, Palmsten K, MacDonald B, Kieran MW, Potenta S, Vong S. Basement membrane derived fibulin-1 and fibulin-5 function as angiogenesis inhibitors and suppress tumor growth. Exp Biol Med (Maywood) 2008; 233(2):155-62. doi: 10.3181/0706-rm-167 [Crossref] [ Google Scholar]
- Senapati S, Rachagani S, Chaudhary K, Johansson SL, Singh RK, Batra SK. Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene 2010; 29(9):1293-302. doi: 10.1038/onc.2009.420 [Crossref] [ Google Scholar]
- Mimeault M, Batra SK. Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J Cell Physiol 2010; 224(3):626-35. doi: 10.1002/jcp.22196 [Crossref] [ Google Scholar]
- Liu T, Bauskin AR, Zaunders J, Brown DA, Pankhurst S, Russell PJ. Macrophage inhibitory cytokine-1 reduces cell adhesion and induces apoptosis in prostate cancer cells. Cancer Res 2003; 63(16):5034-40. [ Google Scholar]
- Siddiqui JA, Pothuraju R, Khan P, Sharma G, Muniyan S, Seshacharyulu P. Pathophysiological role of growth differentiation factor 15 (GDF15) in obesity, cancer, and cachexia. Cytokine Growth Factor Rev 2022; 64:71-83. doi: 10.1016/j.cytogfr.2021.11.002 [Crossref] [ Google Scholar]
- Tan M, Wang Y, Guan K, Sun Y. PTGF-beta, a type beta transforming growth factor (TGF-beta) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-beta signaling pathway. Proc Natl Acad Sci U S A 2000; 97(1):109-14. doi: 10.1073/pnas.97.1.109 [Crossref] [ Google Scholar]
- Baek SJ, Horowitz JM, Eling TE. Molecular cloning and characterization of human nonsteroidal anti-inflammatory drug-activated gene promoter Basal transcription is mediated by Sp1 and Sp3. J Biol Chem 2001; 276(36):33384-92. doi: 10.1074/jbc.M101814200 [Crossref] [ Google Scholar]
- Baek SJ, Kim JS, Moore SM, Lee SH, Martinez J, Eling TE. Cyclooxygenase inhibitors induce the expression of the tumor suppressor gene EGR-1, which results in the up-regulation of NAG-1, an antitumorigenic protein. Mol Pharmacol 2005; 67(2):356-64. doi: 10.1124/mol.104.005108 [Crossref] [ Google Scholar]
- Shim M, Eling TE. Protein kinase C-dependent regulation of NAG-1/placental bone morphogenic protein/MIC-1 expression in LNCaP prostate carcinoma cells. J Biol Chem 2005; 280(19):18636-42. doi: 10.1074/jbc.M414613200 [Crossref] [ Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem 2009; 284(49):33926-38. doi: 10.1074/jbc.M109.023879 [Crossref] [ Google Scholar]
- Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1α enhances hypoxic gene expression and tumor growth. Mol Cell Biol 2010; 30(1):344-53. doi: 10.1128/mcb.00444-09 [Crossref] [ Google Scholar]
- Wang Q, Shen B, Chen L, Zheng P, Feng H, Hao Q. Extracellular calumenin suppresses ERK1/2 signaling and cell migration by protecting fibulin-1 from MMP-13-mediated proteolysis. Oncogene 2015; 34(8):1006-18. doi: 10.1038/onc.2014.52 [Crossref] [ Google Scholar]
- Nasri Nasrabadi P, Nayeri Z, Gharib E, Salmanipour R, Masoomi F, Mahjoubi F. Establishment of a CALU, AURKA, and MCM2 gene panel for discrimination of metastasis from primary colon and lung cancers. PLoS One 2020; 15(5):e0233717. doi: 10.1371/journal.pone.0233717 [Crossref] [ Google Scholar]
- Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: a promising therapeutic target in cancer. Int J Mol Sci 2022; 23(19):11502. doi: 10.3390/ijms231911502 [Crossref] [ Google Scholar]
- Kim T, Croce CM. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med 2023; 55(7):1314-21. doi: 10.1038/s12276-023-01050-9 [Crossref] [ Google Scholar]
- Romano G, Acunzo M, Nana-Sinkam P. microRNAs as novel therapeutics in cancer. Cancers (Basel) 2021; 13(7):1526. doi: 10.3390/cancers13071526 [Crossref] [ Google Scholar]
- Chen SL, Hu D, Chen TZ, Shen SY, Zhao CF, Wang C. Pan-cancer screening and validation of CALU’s role in EMT regulation and tumor microenvironment in triple-negative breast cancer. J Inflamm Res 2024; 17:6743-64. doi: 10.2147/jir.S477846 [Crossref] [ Google Scholar]
- Parnian J, Hoseindokht M, Khademi Z, Moosavi M, Soheili ZS, Samie S. Calumenin knockdown, by intronic artificial microRNA, to improve expression efficiency of the recombinant human coagulation factor IX. Biotechnol Lett 2022; 44(5-6):713-28. doi: 10.1007/s10529-022-03249-8 [Crossref] [ Google Scholar]
- Dhar AK, Robles-Sikisaka R, Saksmerprome V, Lakshman DK. Biology, genome organization, and evolution of parvoviruses in marine shrimp. Adv Virus Res 2014; 89:85-139. doi: 10.1016/b978-0-12-800172-1.00003-3 [Crossref] [ Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25(17):3389-402. doi: 10.1093/nar/25.17.3389 [Crossref] [ Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods 2001; 25(4):402-8. doi: 10.1006/meth.2001.1262 [Crossref] [ Google Scholar]
- Jahangiri B, Soheili ZS, Asadollahi E, Shamsara M, Shariati V, Zomorodipour A. Interleukin-12 inhibits tumor growth and metastasis promoted by tumor-associated mesenchymal stem cells in triple-negative breast cancer. Cell J 2025; 26(9):543-58. doi: 10.22074/cellj.2024.2036513.1634 [Crossref] [ Google Scholar]
- Banin Hirata BK, Oda JM, Losi Guembarovski R, Ariza CB, de Oliveira CE, Watanabe MA. Molecular markers for breast cancer: prediction on tumor behavior. Dis Markers 2014; 2014:513158. doi: 10.1155/2014/513158 [Crossref] [ Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002; 30(9):e36. doi: 10.1093/nar/30.9.e36 [Crossref] [ Google Scholar]
- Gromov P, Gromova I, Bunkenborg J, Cabezon T, Moreira JM, Timmermans-Wielenga V. Up-regulated proteins in the fluid bathing the tumour cell microenvironment as potential serological markers for early detection of cancer of the breast. Mol Oncol 2010; 4(1):65-89. doi: 10.1016/j.molonc.2009.11.003 [Crossref] [ Google Scholar]
- Kreunin P, Urquidi V, Lubman DM, Goodison S. Identification of metastasis-associated proteins in a human tumor metastasis model using the mass-mapping technique. Proteomics 2004; 4(9):2754-65. doi: 10.1002/pmic.200300767 [Crossref] [ Google Scholar]
- Yu CJ, Chang KP, Chang YJ, Hsu CW, Liang Y, Yu JS. Identification of guanylate-binding protein 1 as a potential oral cancer marker involved in cell invasion using omics-based analysis. J Proteome Res 2011; 10(8):3778-88. doi: 10.1021/pr2004133 [Crossref] [ Google Scholar]
- Wu W, Tang X, Hu W, Lotan R, Hong WK, Mao L. Identification and validation of metastasis-associated proteins in head and neck cancer cell lines by two-dimensional electrophoresis and mass spectrometry. Clin Exp Metastasis 2002; 19(4):319-26. doi: 10.1023/a:1015515119300 [Crossref] [ Google Scholar]
- Wadsworth JT, Somers KD, Cazares LH, Malik G, Adam BL, Stack BC Jr. Serum protein profiles to identify head and neck cancer. Clin Cancer Res 2004; 10(5):1625-32. doi: 10.1158/1078-0432.ccr-0297-3 [Crossref] [ Google Scholar]
- Shen C, Hui Z, Wang D, Jiang G, Wang J, Zhang G. Molecular cloning, identification and analysis of lung squamous cell carcinoma-related genes. Lung Cancer 2002; 38(3):235-41. doi: 10.1016/s0169-5002(02)00300-8 [Crossref] [ Google Scholar]
- Li Y, Sun S, Zhang H, Jing Y, Ji X, Wan Q. CALU promotes lung adenocarcinoma progression by enhancing cell proliferation, migration and invasion. Respir Res 2024; 25(1):267. doi: 10.1186/s12931-024-02901-3 [Crossref] [ Google Scholar]
- Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009; 101(10):736-50. doi: 10.1093/jnci/djp082 [Crossref] [ Google Scholar]
- Schwartz AM, Henson DE, Chen D, Rajamarthandan S. Histologic grade remains a prognostic factor for breast cancer regardless of the number of positive lymph nodes and tumor size: a study of 161 708 cases of breast cancer from the SEER Program. Arch Pathol Lab Med 2014; 138(8):1048-52. doi: 10.5858/arpa.2013-0435-OA [Crossref] [ Google Scholar]
- Nishimura R, Osako T, Okumura Y, Hayashi M, Toyozumi Y, Arima N. Ki-67 as a prognostic marker according to breast cancer subtype and a predictor of recurrence time in primary breast cancer. Exp Ther Med 2010; 1(5):747-54. doi: 10.3892/etm.2010.133 [Crossref] [ Google Scholar]
- Eling TE, Baek SJ, Shim M, Lee CH. NSAID activated gene (NAG-1), a modulator of tumorigenesis. J Biochem Mol Biol 2006; 39(6):649-55. doi: 10.5483/bmbrep.2006.39.6.649 [Crossref] [ Google Scholar]
- Emmerson PJ, Duffin KL, Chintharlapalli S, Wu X. GDF15 and growth control. Front Physiol 2018; 9:1712. doi: 10.3389/fphys.2018.01712 [Crossref] [ Google Scholar]
- Spanopoulou A, Gkretsi V. Growth differentiation factor 15 (GDF15) in cancer cell metastasis: from the cells to the patients. Clin Exp Metastasis 2020; 37(4):451-64. doi: 10.1007/s10585-020-10041-3 [Crossref] [ Google Scholar]
- Baek SJ, Eling T. Growth differentiation factor 15 (GDF15): a survival protein with therapeutic potential in metabolic diseases. PharmacolTher 2019; 198:46-58. doi: 10.1016/j.pharmthera.2019.02.008 [Crossref] [ Google Scholar]
- Peake BF, Eze SM, Yang L, Castellino RC, Nahta R. Growth differentiation factor 15 mediates epithelial mesenchymal transition and invasion of breast cancers through IGF-1R-FoxM1 signaling. Oncotarget 2017; 8(55):94393-406. doi: 10.18632/oncotarget.21765 [Crossref] [ Google Scholar]
- Sasahara A, Tominaga K, Nishimura T, Yano M, Kiyokawa E, Noguchi M. An autocrine/paracrine circuit of growth differentiation factor (GDF) 15 has a role for maintenance of breast cancer stem-like cells. Oncotarget 2017; 8(15):24869-81. doi: 10.18632/oncotarget.15276 [Crossref] [ Google Scholar]
- Lim JH, Park JW, Min DS, Chang JS, Lee YH, Park YB. NAG-1 up-regulation mediated by EGR-1 and p53 is critical for quercetin-induced apoptosis in HCT116 colon carcinoma cells. Apoptosis 2007; 12(2):411-21. doi: 10.1007/s10495-006-0576-9 [Crossref] [ Google Scholar]
- Martinez JM, Baek SJ, Mays DM, Tithof PK, Eling TE, Walker NJ. EGR1 is a novel target for AhR agonists in human lung epithelial cells. Toxicol Sci 2004; 82(2):429-35. doi: 10.1093/toxsci/kfh272 [Crossref] [ Google Scholar]
- Cheng JC, Chang HM, Leung PC. Wild-type p53 attenuates cancer cell motility by inducing growth differentiation factor-15 expression. Endocrinology 2011; 152(8):2987-95. doi: 10.1210/en.2011-0059 [Crossref] [ Google Scholar]
- Albertoni M, Shaw PH, Nozaki M, Godard S, Tenan M, Hamou MF. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene 2002; 21(27):4212-9. doi: 10.1038/sj.onc.1205610 [Crossref] [ Google Scholar]
- Zheng P, Wang Q, Teng J, Chen J. Calumenin and fibulin-1 on tumor metastasis: implications for pharmacology. Pharmacol Res 2015; 99:11-5. doi: 10.1016/j.phrs.2015.05.001 [Crossref] [ Google Scholar]
- Tran H, VanDusen WJ, Argraves WS. The self-association and fibronectin-binding sites of fibulin-1 map to calcium-binding epidermal growth factor-like domains. J Biol Chem 1997; 272(36):22600-6. doi: 10.1074/jbc.272.36.22600 [Crossref] [ Google Scholar]
- Johansson S, Svineng G, Wennerberg K, Armulik A, Lohikangas L. Fibronectin-integrin interactions. Front Biosci 1997; 2:d126-46. doi: 10.2741/a178 [Crossref] [ Google Scholar]
- Huang W, Chiquet-Ehrismann R, Moyano JV, Garcia-Pardo A, Orend G. Interference of tenascin-C with syndecan-4 binding to fibronectin blocks cell adhesion and stimulates tumor cell proliferation. Cancer Res 2001; 61(23):8586-94. [ Google Scholar]
- Argraves WS, Dickerson K, Burgess WH, Ruoslahti E. Fibulin, a novel protein that interacts with the fibronectin receptor beta subunit cytoplasmic domain. Cell 1989; 58(4):623-9. doi: 10.1016/0092-8674(89)90097-4 [Crossref] [ Google Scholar]
- Youssef KK, Narwade N, Arcas A, Marquez-Galera A, Jiménez-Castaño R, Lopez-Blau C. Two distinct epithelial-to-mesenchymal transition programs control invasion and inflammation in segregated tumor cell populations. Nat Cancer 2024; 5(11):1660-80. doi: 10.1038/s43018-024-00839-5 [Crossref] [ Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 2011; 27:347-76. doi: 10.1146/annurev-cellbio-092910-154036 [Crossref] [ Google Scholar]
- Zhang Y, Wang X, Zhang M, Zhang Z, Jiang L, Li L. GDF15 promotes epithelial-to-mesenchymal transition in colorectal [corrected]. Artif Cells NanomedBiotechnol 2018; 46(sup2):652-8. doi: 10.1080/21691401.2018.1466146 [Crossref] [ Google Scholar]
- Okamoto M, Koma YI, Kodama T, Nishio M, Shigeoka M, Yokozaki H. Growth differentiation factor 15 promotes progression of esophageal squamous cell carcinoma via TGF-β type II receptor activation. Pathobiology 2020; 87(2):100-13. doi: 10.1159/000504394 [Crossref] [ Google Scholar]
- Georgakopoulos-Soares I, Chartoumpekis DV, Kyriazopoulou V, Zaravinos A. EMT factors and metabolic pathways in cancer. Front Oncol 2020; 10:499. doi: 10.3389/fonc.2020.00499 [Crossref] [ Google Scholar]
- Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016; 166(1):21-45. doi: 10.1016/j.cell.2016.06.028 [Crossref] [ Google Scholar]
- Ludvigsen M, Thorlacius-Ussing L, Vorum H, Moyer MP, Stender MT, Thorlacius-Ussing O. Proteomic characterization of colorectal cancer cells versus normal-derived colon mucosa cells: approaching identification of novel diagnostic protein biomarkers in colorectal cancer. Int J Mol Sci 2020; 21(10):3466. doi: 10.3390/ijms21103466 [Crossref] [ Google Scholar]
- Zhang M, Cao C, Li X, Gu Q, Xu Y, Zhu Z. Five EMT-related genes signature predicts overall survival and immune environment in microsatellite instability-high gastric cancer. Cancer Med 2023; 12(2):2075-88. doi: 10.1002/cam4.4975 [Crossref] [ Google Scholar]
- Liu K, Liu PC, Liu R, Wu X. Dual AO/EB staining to detect apoptosis in osteosarcoma cells compared with flow cytometry. Med Sci Monit Basic Res 2015; 21:15-20. doi: 10.12659/msmbr.893327 [Crossref] [ Google Scholar]
- Wang Y, Sun Y, Fu Y, Guo X, Long J, Xuan LY. Calumenin relieves cardiac injury by inhibiting ERS-initiated apoptosis during viral myocarditis. Int J Clin Exp Pathol 2017; 10(7):7277-84. [ Google Scholar]
- Tang H, Ma M, Dai J, Cui C, Si L, Sheng X. miR-let-7b and miR-let-7c suppress tumourigenesis of human mucosal melanoma and enhance the sensitivity to chemotherapy. J Exp Clin Cancer Res 2019; 38(1):212. doi: 10.1186/s13046-019-1190-3 [Crossref] [ Google Scholar]
- Fultang N, Chakraborty M, Peethambaran B. Regulation of cancer stem cells in triple negative breast cancer. Cancer Drug Resist 2021; 4(2):321-42. doi: 10.20517/cdr.2020.106 [Crossref] [ Google Scholar]
- Nagano K, Imai S, Zhao X, Yamashita T, Yoshioka Y, Abe Y. Identification and evaluation of metastasis-related proteins, oxysterol binding protein-like 5 and calumenin, in lung tumors. Int J Oncol 2015; 47(1):195-203. doi: 10.3892/ijo.2015.3000 [Crossref] [ Google Scholar]
- Jung DH, Mo SH, Kim DH. Calumenin, a multiple EF-hands Ca2 + -binding protein, interacts with ryanodine receptor-1 in rabbit skeletal sarcoplasmic reticulum. BiochemBiophys Res Commun 2006; 343(1):34-42. doi: 10.1016/j.bbrc.2006.02.115 [Crossref] [ Google Scholar]
- Sahoo SK, Kim DH. Calumenin interacts with SERCA2 in rat cardiac sarcoplasmic reticulum. Mol Cells 2008; 26(3):265-9. doi: 10.1016/s1016-8478(23)13994-x [Crossref] [ Google Scholar]
- Sahoo SK, Kim DH. Characterization of calumenin in mouse heart. BMB Rep 2010; 43(3):158-63. doi: 10.5483/bmbrep.2010.43.3.158 [Crossref] [ Google Scholar]
- Lee JH, Kwon EJ, Kim DH. Calumenin has a role in the alleviation of ER stress in neonatal rat cardiomyocytes. BiochemBiophys Res Commun 2013; 439(3):327-32. doi: 10.1016/j.bbrc.2013.08.087 [Crossref] [ Google Scholar]
- Pan ST, Li ZL, He ZX, Qiu JX, Zhou SF. Molecular mechanisms for tumour resistance to chemotherapy. Clin Exp PharmacolPhysiol 2016; 43(8):723-37. doi: 10.1111/1440-1681.12581 [Crossref] [ Google Scholar]
- Gil D, Ciołczyk-Wierzbicka D, Dulińska-Litewka J, Zwawa K, McCubrey JA, Laidler P. The mechanism of contribution of integrin linked kinase (ILK) to epithelial-mesenchymal transition (EMT). Adv Enzyme Regul 2011; 51(1):195-207. doi: 10.1016/j.advenzreg.2010.09.005 [Crossref] [ Google Scholar]
- Brandão-Costa RM, Helal-Neto E, Vieira AM, Barcellos-de-Souza P, Morgado-Diaz J, Barja-Fidalgo C. Extracellular matrix derived from high metastatic human breast cancer triggers epithelial-mesenchymal transition in epithelial breast cancer cells through αvβ3 integrin. Int J Mol Sci 2020; 21(8):2995. doi: 10.3390/ijms21082995 [Crossref] [ Google Scholar]
- Donnelly SM, Paplomata E, Peake BM, Sanabria E, Chen Z, Nahta R. P38 MAPK contributes to resistance and invasiveness of HER2- overexpressing breast cancer. Curr Med Chem 2014; 21(4):501-10. doi: 10.2174/0929867320666131119155023 [Crossref] [ Google Scholar]
- Zhao X, Liu X, Hu S, Pan Y, Zhang J, Tai G. GDF15 contributes to radioresistance by mediating the EMT and stemness of breast cancer cells. Int J Mol Sci 2022; 23(18):10911. doi: 10.3390/ijms231810911 [Crossref] [ Google Scholar]
- Li S, Ma YM, Zheng PS, Zhang P. GDF15 promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp Clin Cancer Res 2018; 37(1):80. doi: 10.1186/s13046-018-0744-0 [Crossref] [ Google Scholar]
- Rzemieniewski B, Kasztelan A, Poboży K, Domańska-Poboża J. Growth differentiation factor 15 - a review of current literature on biological roles and clinical significance. Eur J Clin Exp Med 2024; 22(4):921-33. doi: 10.15584/ejcem.2024.4.19 [Crossref] [ Google Scholar]
- Menon R, Roy A, Mukherjee S, Belkin S, Zhang Y, Omenn GS. Functional implications of structural predictions for alternative splice proteins expressed in Her2/neu-induced breast cancers. J Proteome Res 2011; 10(12):5503-11. doi: 10.1021/pr200772w [Crossref] [ Google Scholar]
- Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNA therapeutics in cancer - an emerging concept. EBioMedicine 2016; 12:34-42. doi: 10.1016/j.ebiom.2016.09.017 [Crossref] [ Google Scholar]
- Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids 2015; 4(9):e252. doi: 10.1038/mtna.2015.23 [Crossref] [ Google Scholar]